4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | GLYCEROL, MGS-M2 | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | 登録日 | 2012-11-26 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3N
 
 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3M
 
 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | MGS-M4, SODIUM ION, SULFATE ION | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
1F9X
 
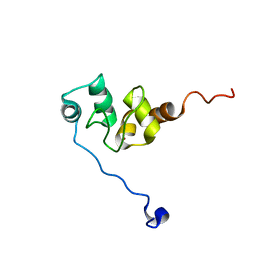 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | 分子名称: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | 著者 | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | 登録日 | 2000-07-11 | | 公開日 | 2001-07-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
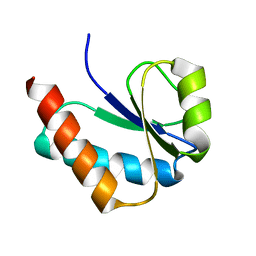
1VKR
 
 | |
1VRV
 
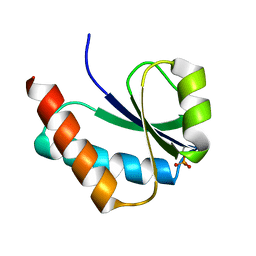 | |
1ZHS
 
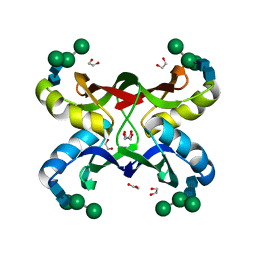 | | Crystal structure of MVL bound to Man3GlcNAc2 | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | 登録日 | 2005-04-26 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
2WY2
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | 分子名称: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | 著者 | Sang, Y.S, Cai, M, Clore, G.M. | | 登録日 | 2009-11-11 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
6FCE
 
 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | 分子名称: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | 著者 | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | 登録日 | 2017-12-20 | | 公開日 | 2018-04-25 | | 最終更新日 | 2018-05-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | 分子名称: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | 著者 | Sang, Y.S, Cai, M, Clore, G.M. | | 登録日 | 2009-10-29 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
1ZHQ
 
 | | Crystal structure of apo MVL | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, mannan-binding lectin | | 著者 | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | 登録日 | 2005-04-26 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
2LRL
 
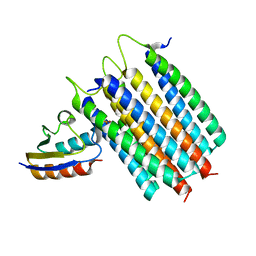 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System | | 分子名称: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphocarrier protein HPr | | 著者 | Jung, Y, Cai, M, Clore, M. | | 登録日 | 2012-04-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
1W8H
 
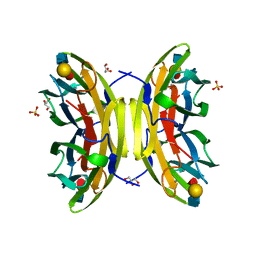 | | structure of pseudomonas aeruginosa lectin II (PA-IIL)complexed with lewisA trisaccharide | | 分子名称: | CALCIUM ION, GLYCEROL, PSEUDOMONAS AERUGINOSA LECTIN II, ... | | 著者 | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | 登録日 | 2004-09-21 | | 公開日 | 2005-03-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1W8F
 
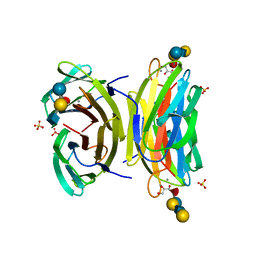 | | PSEUDOMONAS AERUGINOSA LECTIN II (PA-IIL)COMPLEXED WITH LACTO-N-NEO- FUCOPENTAOSE V(LNPFV) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | 登録日 | 2004-09-21 | | 公開日 | 2005-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
4JLV
 
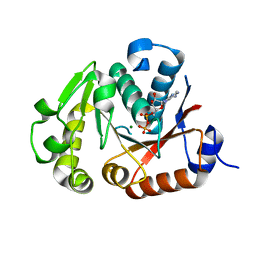 | | Crystal structure of the chimerical protein CapA1B1 in complex with ADP-Mg | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, C-terminal fragment of Membrane protein CapA1, Putative uncharacterized protein capB1, ... | | 著者 | Gruszczyk, J, Olivares-Illana, V, Nourikyan, J, Fleurie, A, Bechet, E, Aumont-Nicaise, M, Gueguen-Chaignon, V, Morera, S, Grangeasse, C, Nessler, S. | | 登録日 | 2013-03-13 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus
Plos One, 8, 2013
|
|
1P9P
 
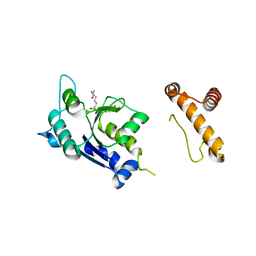 | | The Crystal Structure of a M1G37 tRNA Methyltransferase, TrmD | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | 著者 | Elkins, P.A, Watts, J.M, Zalacain, M, Van Thiel, A, Vitaszka, P.R, Redlak, M, Andraos-Selim, C, Rastinejad, F, Holmes, W.M. | | 登録日 | 2003-05-12 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
J.Mol.Biol., 333, 2003
|
|
3P27
 
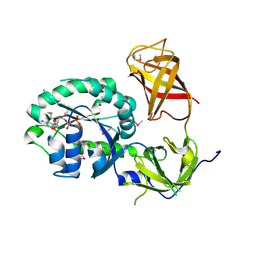 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | 分子名称: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | 著者 | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | 登録日 | 2010-10-01 | | 公開日 | 2010-11-17 | | 最終更新日 | 2012-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P26
 
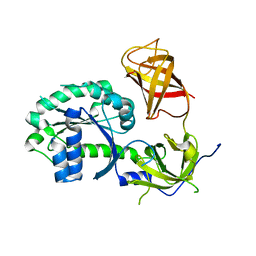 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | 分子名称: | Elongation factor 1 alpha-like protein | | 著者 | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | 登録日 | 2010-10-01 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4GPK
 
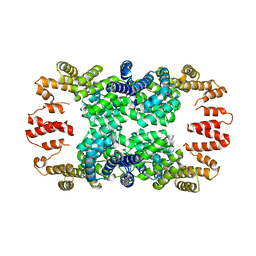 | | Crystal structure of NprR in complex with its cognate peptide NprX | | 分子名称: | NprR, NprX peptide | | 著者 | Zouhir, S, Guimaraes, B, Perchat, S, Nicaise, M, Lereclus, D, Nessler, S. | | 登録日 | 2012-08-21 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Nucleic Acids Res., 41, 2013
|
|
7NA2
 
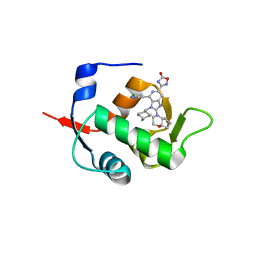 | | HDM2 in complex with compound 56 | | 分子名称: | 3-[4-(5-chloropyridin-3-yl)-2-[(4aR,7aR)-hexahydrocyclopenta[b][1,4]oxazin-4(4aH)-yl]-3-{[(1r,4R)-4-methylcyclohexyl]methyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2 | | 著者 | Scapin, G. | | 登録日 | 2021-06-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA3
 
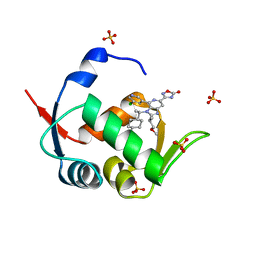 | | HDM2 in complex with compound 62 | | 分子名称: | 3-[4-(5-chloropyridin-3-yl)-2-[(2S)-1-methoxypropan-2-yl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Scapin, G. | | 登録日 | 2021-06-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA1
 
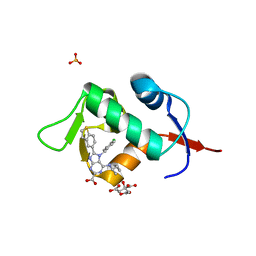 | | HDM2 in complex with compound 2 | | 分子名称: | 8-(1-benzothiophen-5-yl)-7-[(4-chlorophenyl)methyl]-6-{[(1R)-1-cyclopropylethyl]amino}-7H-purine-2-carboxylic acid, CITRIC ACID, E3 ubiquitin-protein ligase Mdm2, ... | | 著者 | Scapin, G. | | 登録日 | 2021-06-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA4
 
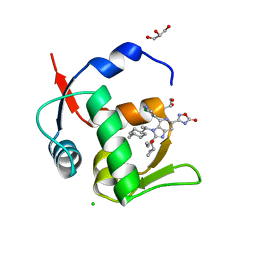 | | HDM2 in complex with compound 63 | | 分子名称: | 3-[4-(5-chloropyridin-3-yl)-2-[(R)-cyclopropyl(ethoxy)methyl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, CHLORIDE ION, GLYCEROL, ... | | 著者 | Scapin, G. | | 登録日 | 2021-06-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
