2M7Q
 
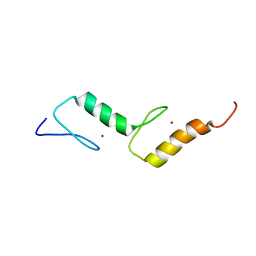 | | Solution structure of TAX1BP1 UBZ1+2 | | 分子名称: | Tax1-binding protein 1, ZINC ION | | 著者 | Ceregido, M.A, Spinola Amilibia, M, Buts, L, Bravo, J, van Nuland, N.A.J. | | 登録日 | 2013-04-29 | | 公開日 | 2013-12-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of TAX1BP1 UBZ1+2 provides insight into target specificity and adaptability.
J.Mol.Biol., 426, 2014
|
|
5CXC
 
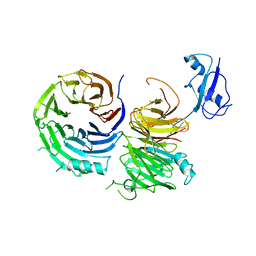 | |
5CYK
 
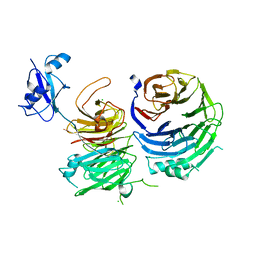 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | 分子名称: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | 著者 | Wegrecki, M, Bravo, J. | | 登録日 | 2015-07-30 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5CXB
 
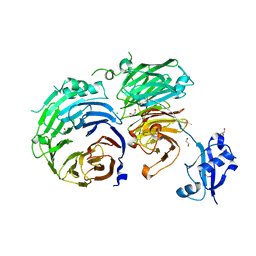 | |
6EXX
 
 | |
6EZ7
 
 | | Pes4 RRM3 Structure | | 分子名称: | DI(HYDROXYETHYL)ETHER, Protein PES4 | | 著者 | Mohamad, N, Bravo, J. | | 登録日 | 2017-11-14 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pes4 RRM3 Structure
To Be Published
|
|
5D77
 
 | |
5D78
 
 | |
1E50
 
 | | AML1/CBFbeta complex | | 分子名称: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | 著者 | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | 登録日 | 2000-07-13 | | 公開日 | 2001-07-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
1B7B
 
 | | Carbamate kinase from Enterococcus faecalis | | 分子名称: | CARBAMATE KINASE, SULFATE ION | | 著者 | Marina, A, Alzari, P.M, Bravo, J, Uriarte, M, Barcelona, B, Fita, I, Rubio, V. | | 登録日 | 1999-01-20 | | 公開日 | 2000-01-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Carbamate kinase: New structural machinery for making carbamoyl phosphate, the common precursor of pyrimidines and arginine.
Protein Sci., 8, 1999
|
|
2J6O
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETEROTRIMER | | 分子名称: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | 著者 | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | 登録日 | 2006-10-02 | | 公開日 | 2006-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6F
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) BOUND TO CBL-B PEPTIDE | | 分子名称: | CD2-ASSOCIATED PROTEIN, E3 UBIQUITIN-PROTEIN LIGASE CBL-B | | 著者 | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | 登録日 | 2006-09-28 | | 公開日 | 2006-10-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6K
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) | | 分子名称: | CD2-ASSOCIATED PROTEIN, SODIUM ION | | 著者 | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | 登録日 | 2006-09-29 | | 公開日 | 2006-10-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J7I
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETERODIMER | | 分子名称: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | 著者 | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | 登録日 | 2006-10-09 | | 公開日 | 2006-11-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Atypical Polyproline Recognition by the Cms N-Terminal Src Homology 3 Domain.
J.Biol.Chem., 281, 2006
|
|
2IWL
 
 | |
1EVS
 
 | | CRYSTAL STRUCTURE OF HUMAN ONCOSTATIN M | | 分子名称: | ONCOSTATIN M | | 著者 | Deller, M.C, Hudson, K.R, Ikemizu, S, Bravo, J, Jones, E.Y, Heath, J.K. | | 登録日 | 2000-04-20 | | 公開日 | 2000-09-13 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and functional dissection of the cytostatic cytokine oncostatin M.
Structure Fold.Des., 8, 2000
|
|
1QF7
 
 | |
1CF9
 
 | |
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | 著者 | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | 登録日 | 2023-06-06 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | 著者 | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
2MCN
 
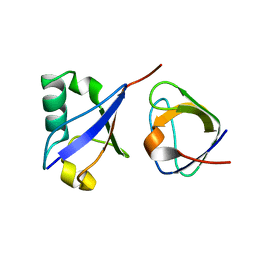 | | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications | | 分子名称: | CD2-associated protein, Ubiquitin | | 著者 | Ortega-Roldan, J, Salmon, L, Azuaga, A, Blackledge, M, Van Nuland, N. | | 登録日 | 2013-08-22 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications.
Plos One, 8, 2013
|
|
2LZ6
 
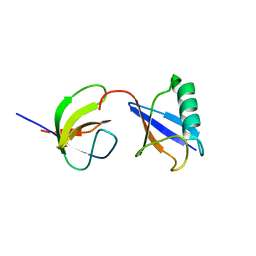 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | 分子名称: | CD2-associated protein, Ubiquitin | | 著者 | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | 登録日 | 2012-09-24 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
6JQQ
 
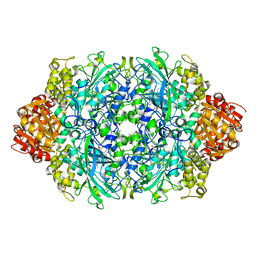 | |
2WE5
 
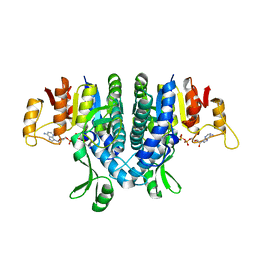 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | 著者 | Ramon-Maiques, S, Marina, A, Rubio, V. | | 登録日 | 2009-03-27 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2WE4
 
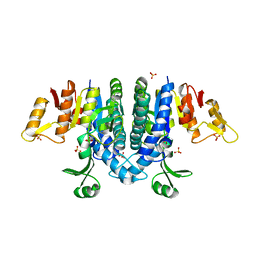 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | 分子名称: | CARBAMATE KINASE 1, SULFATE ION | | 著者 | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | 登録日 | 2009-03-27 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
