6Y5A
 
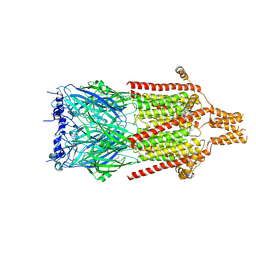 | | Serotonin-bound 5-HT3A receptor in Salipro | | 分子名称: | 5-hydroxytryptamine receptor 3A, SEROTONIN | | 著者 | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | 登録日 | 2020-02-25 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
5M5G
 
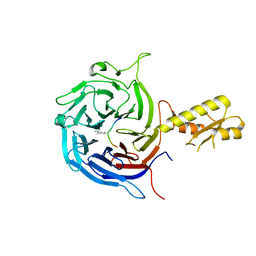 | | Crystal structure of the Chaetomium Thermophilum polycomb repressive complex 2 (PRC2) | | 分子名称: | Fragment from molecular 2 (region containing putative polycomb protein Suz12), HISTONE H3 11-Mer peptide, Putative uncharacterized protein, ... | | 著者 | Zhang, Y, Justin, N, Wilson, J, Gamblin, S. | | 登録日 | 2016-10-21 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
7ST4
 
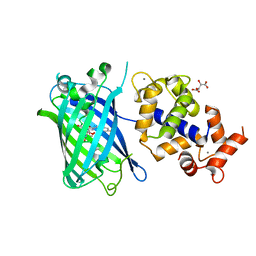 | | Calcium-saturated jGCaMP8.410.80 | | 分子名称: | CALCIUM ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Zhang, Y, Looger, L.L. | | 登録日 | 2021-11-11 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fast and sensitive GCaMP calcium indicators for imaging neural populations
Nature, 615, 2023
|
|
1FKC
 
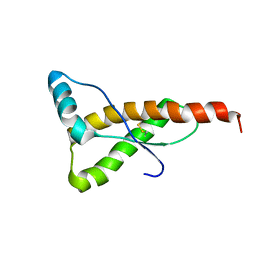 | | HUMAN PRION PROTEIN (MUTANT E200K) FRAGMENT 90-231 | | 分子名称: | PRION PROTEIN | | 著者 | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | 登録日 | 2000-08-09 | | 公開日 | 2000-09-21 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
1FO7
 
 | | HUMAN PRION PROTEIN MUTANT E200K FRAGMENT 90-231 | | 分子名称: | PRION PROTEIN | | 著者 | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | 登録日 | 2000-08-25 | | 公開日 | 2000-09-21 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
2R84
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP and AICAR | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | 著者 | Zhang, Y, White, R.H, Ealick, S.E. | | 登録日 | 2007-09-10 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | 分子名称: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | 著者 | Zhang, Y, White, R.H, Ealick, S.E. | | 登録日 | 2007-09-09 | | 公開日 | 2007-12-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7N
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | 分子名称: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, White, R.H, Ealick, S.E. | | 登録日 | 2007-09-09 | | 公開日 | 2007-12-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R87
 
 | |
1N6G
 
 | | The structure of immature Dengue-2 prM particles | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-10 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
 | | The structure of immature Yellow Fever virus particle | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-26 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
5T0X
 
 | |
6O3W
 
 | |
6O3X
 
 | |
3OMX
 
 | | Crystal structure of Ssu72 with vanadate complex | | 分子名称: | CG14216, VANADATE ION | | 著者 | Zhang, Y, Zhang, M, Zhang, Y. | | 登録日 | 2010-08-27 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3366 Å) | | 主引用文献 | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | 分子名称: | CG14216 | | 著者 | Zhang, Y, Zhang, M, Zhang, Y. | | 登録日 | 2010-08-27 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8701 Å) | | 主引用文献 | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
5E00
 
 | | Structure of HLA-A2 P130 | | 分子名称: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | 著者 | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | 登録日 | 2015-09-26 | | 公開日 | 2017-01-18 | | 最終更新日 | 2019-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | 分子名称: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-22 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1THD
 
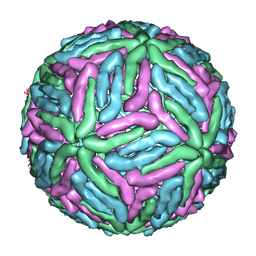 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | 分子名称: | Major envelope protein E | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2004-06-01 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
4KM9
 
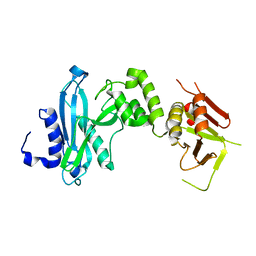 | |
1O7O
 
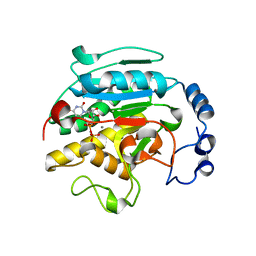 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
4KM8
 
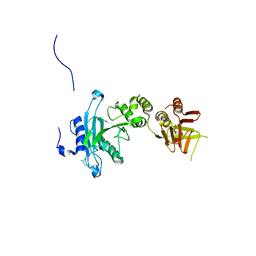 | | Crystal structure of Sufud60 | | 分子名称: | Suppressor of fused homolog | | 著者 | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | 登録日 | 2013-05-08 | | 公開日 | 2013-11-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-12 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
