3IYK
 
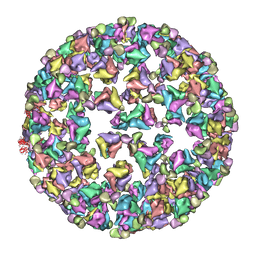 | | Bluetongue virus structure reveals a sialic acid binding domain, amphipathic helices and a central coiled coil in the outer capsid proteins | | 分子名称: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, VP2, VP5 | | 著者 | Zhang, X, Boyce, M, Bhattacharya, B, Zhang, X, Schein, S, Roy, P, Zhou, Z.H. | | 登録日 | 2010-01-25 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1ML9
 
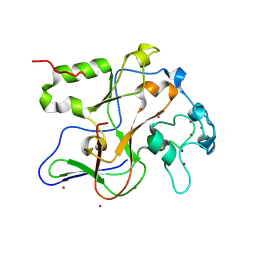 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | 分子名称: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | 著者 | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | 登録日 | 2002-08-30 | | 公開日 | 2002-10-23 | | 最終更新日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
1ORI
 
 | |
1OR8
 
 | |
1ORH
 
 | |
4Y4M
 
 | | Thiazole synthase Thi4 from Methanocaldococcus jannaschii | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Putative ribose 1,5-bisphosphate isomerase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R)-2,3,5-tris(oxidanyl)-4-oxidanylidene-pentyl] hydrogen phosphate | | 著者 | Zhang, X, Ealick, S.E. | | 登録日 | 2015-02-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
4Y4L
 
 | | Crystal structure of yeast Thi4-C205S | | 分子名称: | (2E)-2-[(2S,4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-yl]iminoethanoic acid, Thiamine thiazole synthase | | 著者 | Zhang, X, Ealick, S.E. | | 登録日 | 2015-02-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
2B5Y
 
 | |
2B5X
 
 | |
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | 分子名称: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | 著者 | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | 登録日 | 2003-05-21 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
3KL1
 
 | |
1HQK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | 分子名称: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | 著者 | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | 登録日 | 2000-12-18 | | 公開日 | 2001-12-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
3KLX
 
 | |
4KSY
 
 | |
4O67
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with GAMP | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.444 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O68
 
 | | Structure of human cyclic GMP-AMP synthase (cGAS) | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.436 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O6A
 
 | | Mouse cyclic GMP-AMP synthase (cGAS) in complex with DNA | | 分子名称: | Cyclic GMP-AMP synthase, DNA1, DNA2, ... | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O69
 
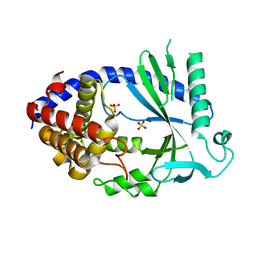 | | Human cyclic GMP-AMP synthase (cGAS) in complex with sulfate ion | | 分子名称: | Cyclic GMP-AMP synthase, SULFATE ION, ZINC ION | | 著者 | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
1EUB
 
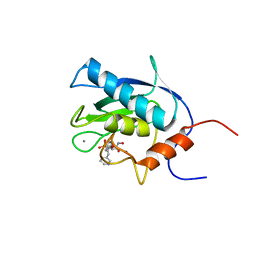 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | 分子名称: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | 著者 | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | 登録日 | 2000-04-14 | | 公開日 | 2001-04-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
1CDZ
 
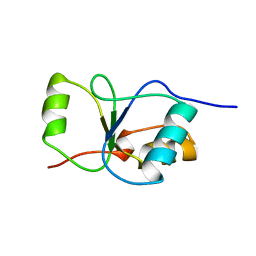 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | 分子名称: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | 著者 | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | 登録日 | 1999-03-04 | | 公開日 | 2000-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
1CFA
 
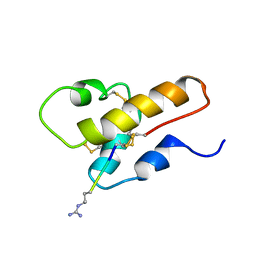 | | SOLUTION STRUCTURE OF A SEMI-SYNTHETIC C5A RECEPTOR ANTAGONIST AT PH 5.2, 303K, NMR, 20 STRUCTURES | | 分子名称: | COMPLEMENT 5A SEMI-SYNTHETIC ANTAGONIST, SYNTHETIC N-TERMINAL TAIL | | 著者 | Zhang, X, Boyar, W, Galakatos, N, Gonnella, N.C. | | 登録日 | 1996-09-21 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding.
Protein Sci., 6, 1997
|
|
1E32
 
 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, P97 | | 著者 | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | 登録日 | 2000-06-05 | | 公開日 | 2001-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
1UON
 
 | | REOVIRUS POLYMERASE LAMBDA-3 LOCALIZED BY ELECTRON CRYOMICROSCOPY OF VIRIONS AT 7.6-A RESOLUTION | | 分子名称: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*GP*GP*GP*GP*GP*)-3', 5'-R(*UP*AP*GP*CP*CP*CP*CP*CP*)-3', ... | | 著者 | Zhang, X, Walker, S.B, Chipman, P.R, Nibert, M.L, Baker, T.S. | | 登録日 | 2003-09-21 | | 公開日 | 2003-11-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Reovirus Polymerase Lambda3 Localized by Cryo-Electron Microscopy of Virions at a Resolution of 7.6 A
Nat.Struct.Biol., 10, 2003
|
|
5HWL
 
 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | 分子名称: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | 著者 | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | 登録日 | 2016-01-29 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | 著者 | Zhang, X, Zhang, Q, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
