7CZW
 
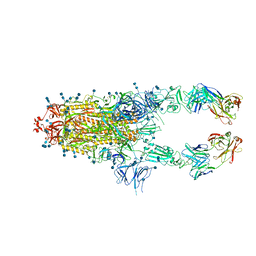 | | S protein of SARS-CoV-2 in complex bound with P5A-2G7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0D
 
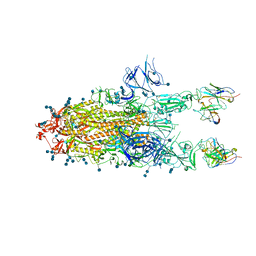 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | 著者 | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
6R46
 
 | |
6TKZ
 
 | |
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.97 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8JPP
 
 | | Cryo-EM structure of succinate receptor bound to succinate acid coupling MiniGsq | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | 登録日 | 2023-06-12 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 2024
|
|
8Q84
 
 | |
5YSL
 
 | | Crystal structure of antibody 1H1 Fab | | 分子名称: | 1H1 heavy chain, 1H1 light chain | | 著者 | Hu, X.L, Yang, F.L. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design.
PLoS Pathog., 13, 2017
|
|
1XHO
 
 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | 分子名称: | Chorismate mutase, UNKNOWN ATOM OR ION | | 著者 | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-09-20 | | 公開日 | 2004-11-23 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6PXE
 
 | |
5GTU
 
 | |
8JAT
 
 | |
5YFG
 
 | |
6PSK
 
 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | 著者 | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | 登録日 | 2019-07-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2020-08-12 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
7XGR
 
 | |
8J5X
 
 | | The crystal structure of TrkA(G595R) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.09192252 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J5W
 
 | | The crystal structure of TrkA(F589L) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.28041458 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8XAB
 
 | |
7XDT
 
 | |
8J61
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.05065274 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J63
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | 分子名称: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | 著者 | Zhang, Z.M, Wang, Y.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.0005 Å) | | 主引用文献 | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | 分子名称: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | 著者 | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | 登録日 | 2016-08-08 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5NHF
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-21 | | 公開日 | 2017-04-19 | | 最終更新日 | 2017-05-10 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | 分子名称: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | 著者 | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | 登録日 | 2016-08-08 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5NHJ
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-21 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
