7MKI
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XWW
 
 | | Crystal structure of NTR in complex with BN-XB | | 分子名称: | 2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-8-[1-[(4-nitrophenyl)methyl]pyridin-1-ium-4-yl]-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaene, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Chen, X, Chen, J, Li, J.L. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of NTR in complex with BN-XB
To Be Published
|
|
3V16
 
 | | An intramolecular pi-cation latch in phosphatidylinositol-specific phospholipase C from S.aureus controls substrate access to the active site | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, CHLORIDE ION | | 著者 | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | 登録日 | 2011-12-09 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
3V18
 
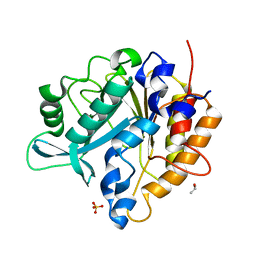 | | Structure of the Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | 分子名称: | 1-phosphatidylinositol phosphodiesterase, ISOPROPYL ALCOHOL, SULFATE ION | | 著者 | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | 登録日 | 2011-12-09 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
6R6P
 
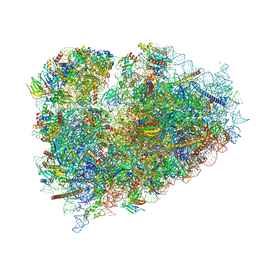 | | Structure of XBP1u-paused ribosome nascent chain complex (rotated state) | | 分子名称: | 18S rRNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
5W81
 
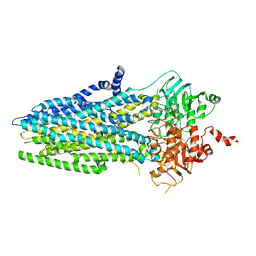 | | Phosphorylated, ATP-bound structure of zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | 著者 | Zhang, Z, Liu, F, Chen, J. | | 登録日 | 2017-06-21 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Conformational Changes of CFTR upon Phosphorylation and ATP Binding.
Cell, 170, 2017
|
|
7L7B
 
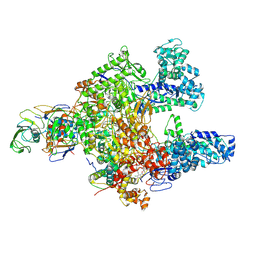 | | Clostridioides difficile RNAP with fidaxomicin | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Boyaci, H, Campbell, E.A, Darst, S.A, Chen, J. | | 登録日 | 2020-12-28 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile.
Nature, 604, 2022
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | 著者 | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | 登録日 | 2021-11-18 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VZF
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-11-16 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
7YDL
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | 登録日 | 2022-07-04 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
8K2A
 
 | | Cryo-EM structure of the human 55S mitoribosome with Tigecycline | | 分子名称: | 12S rRNA, 16S rRNA, 39S ribosomal protein L22, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2B
 
 | | Cryo-EM structure of the human 39S mitoribosome with Tigecycline | | 分子名称: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2D
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | 分子名称: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Buschauer, R, Beckmann, R, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K82
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, Not5 and P-site tRNA | | 分子名称: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Buschauer, R, Beckmann, R, Cheng, J. | | 登録日 | 2023-07-28 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | 分子名称: | Major prion protein | | 著者 | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-01-18 | | 公開日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
8PVL
 
 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV8
 
 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV2
 
 | | Chaetomium thermophilum pre-60S State 10 - pre-5S rotation with Ytm1-Erb1 | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PUW
 
 | | Chaetomium thermophilum Las1-Grc3-complex | | 分子名称: | Las1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV3
 
 | | Chaetomium thermophilum pre-60S State 9 - pre-5S rotation - immature H68/H69 - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PTW
 
 | | Chaetomium thermophilum Rix1-complex | | 分子名称: | Pre-rRNA-processing protein IPI3, Pre-rRNA-processing protein RIX1 | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-15 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV7
 
 | | Chaetomium thermophilum pre-60S State 1 - pre-5S rotation (Arx1/Nog2 state) - Composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.12 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
