7YOO
 
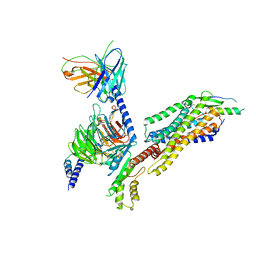 | | Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Kang, H, Park, C, Kim, J, Choi, H.-J. | | 登録日 | 2022-08-01 | | 公開日 | 2023-03-22 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
8SXS
 
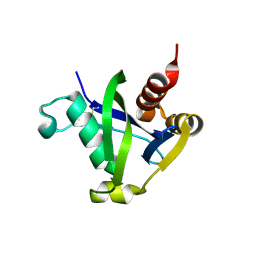 | |
8D86
 
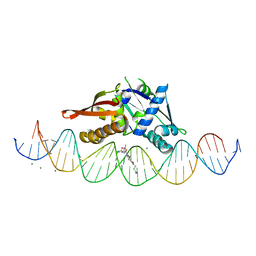 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence co-crystallized with a guest small molecule, netropsin. | | 分子名称: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | 著者 | Orun, A.R, Snow, C.D. | | 登録日 | 2022-06-07 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
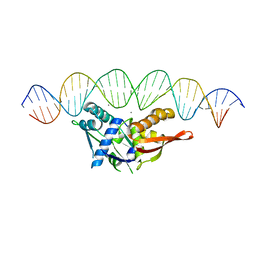
8D8M
 
 | |
8TBW
 
 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) peptide KLSHQPVLL | | 分子名称: | Beta-2-microglobulin, HLA-A*02:01 alpha chain, NITRATE ION, ... | | 著者 | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | 登録日 | 2023-06-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TSY
 
 | |
8TSZ
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | 分子名称: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | 著者 | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | 登録日 | 2023-08-12 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | 分子名称: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | 著者 | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | 登録日 | 2023-08-12 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT0
 
 | |
8TSU
 
 | |
8TT1
 
 | |
8V1J
 
 | |
7KWO
 
 | | rFVIIIFc-VWF-XTEN (BIVV001) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | 著者 | Fuller, J.R, Batchelor, J.D. | | 登録日 | 2020-12-01 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy.
Blood, 137, 2021
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8C3O
 
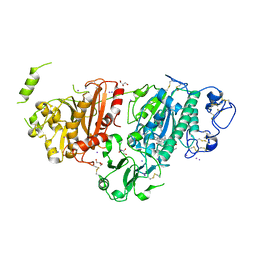 | | Crystal structure of autotaxin gamma and compound MEY-003 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | 著者 | Eymery, M.C, McCarthy, A.A. | | 登録日 | 2022-12-27 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
6TYF
 
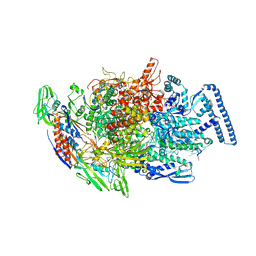 | |
6TYG
 
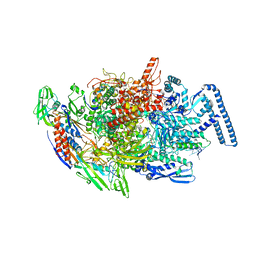 | |
6U0O
 
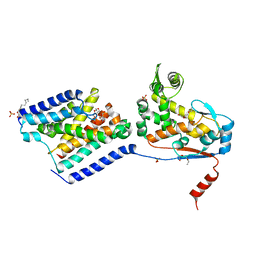 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | 著者 | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | 登録日 | 2019-08-14 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
6TYE
 
 | |
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | 分子名称: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | 著者 | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | 登録日 | 2019-10-29 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
7QYH
 
 | |
6UTM
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | 分子名称: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | 著者 | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | 登録日 | 2019-10-29 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | 分子名称: | GLYCEROL, Lon211 | | 著者 | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | 登録日 | 2019-12-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | 分子名称: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | 著者 | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | 登録日 | 2019-10-29 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
