4MSD
 
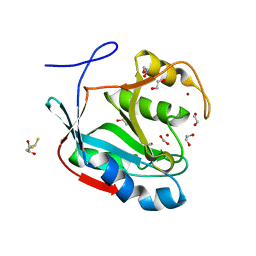 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protein SST2 T319I mutant | | 分子名称: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AMSH-like protease sst2, ... | | 著者 | Shrestha, R.K, Ronau, J.A, Das, C. | | 登録日 | 2013-09-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-08-31 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5T57
 
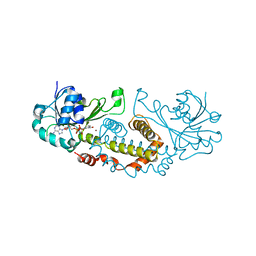 | | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD | | 分子名称: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Semialdehyde dehydrogenase NAD-binding protein, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD
To be published
|
|
4MNI
 
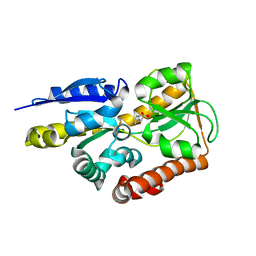 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | 分子名称: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-09-10 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | 分子名称: | HYPOTHETICAL PROTEIN XCC279 | | 著者 | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-10-22 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4MZL
 
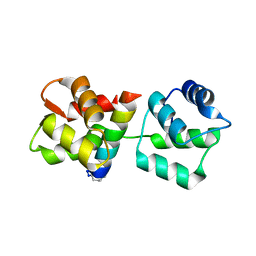 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with HBS myoA, a hydrogen bond surrogate myoA helix mimetic | | 分子名称: | Myosin A tail domain interacting protein, hydrogen bond surrogate (HBS) myoA helix mimetic | | 著者 | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | 登録日 | 2013-09-30 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
4L9W
 
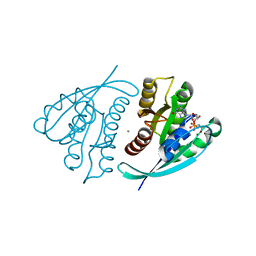 | | Crystal Structure of H-Ras G12C, GMPPNP-bound | | 分子名称: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | 登録日 | 2013-06-18 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
5TMZ
 
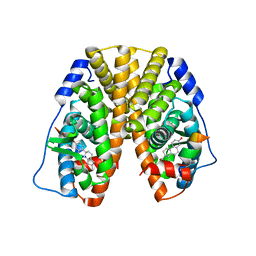 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,17S)-16-(3-methoxybenzyl)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthrene-3,17-diol | | 分子名称: | (9beta,13alpha,14beta,16alpha,17alpha)-16-[(4-methoxyphenyl)methyl]estra-1,3,5(10)-triene-3,17-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.207 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4L47
 
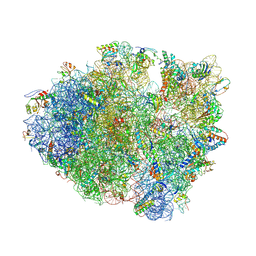 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U on the Ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | 登録日 | 2013-06-07 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.220001 Å) | | 主引用文献 | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5TN7
 
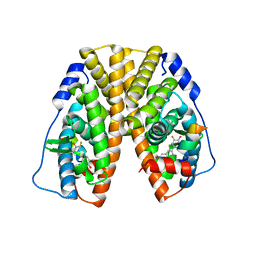 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-3'-fluoro-4'-hydroxy-3-((hydroxyiminio)methyl)-[1,1'-biphenyl]-4-olate | | 分子名称: | 3-fluoro-3'-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Minutolo, F, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.238 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4LEB
 
 | |
4KVD
 
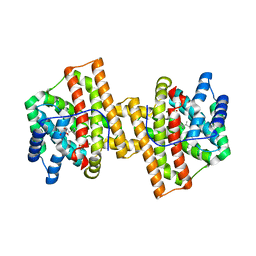 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (4aS,7S)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolin-1-ium | | 分子名称: | (1R,4aS,7S,8aR)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolinium, Aristolochene synthase, GLYCEROL, ... | | 著者 | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4L71
 
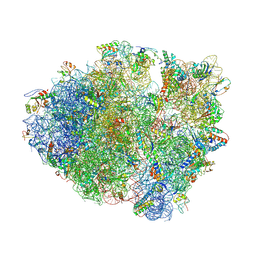 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | 登録日 | 2013-06-13 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.900001 Å) | | 主引用文献 | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5TLU
 
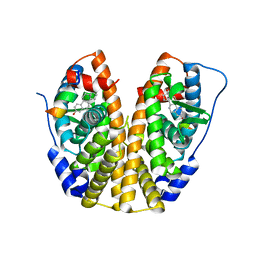 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the PEG-linked Dimeric Estrogen, EE2-(eg)6-EE2-amine | | 分子名称: | (14beta,17alpha)-21-(4-aminophenyl)-19-norpregna-1(10),2,4-trien-20-yne-3,17-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.223 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM3
 
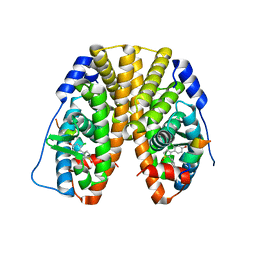 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,3-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | 分子名称: | (1S)-2,3-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TML
 
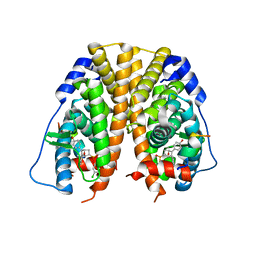 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, (E)-6-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | 分子名称: | 6-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMV
 
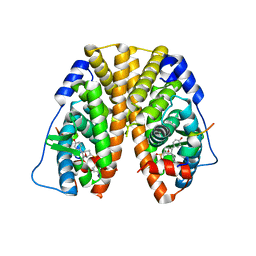 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS analog, 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | 分子名称: | 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN5
 
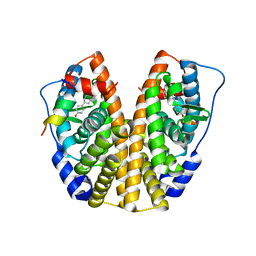 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the AC-ring estrogen, (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | 分子名称: | (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4L8E
 
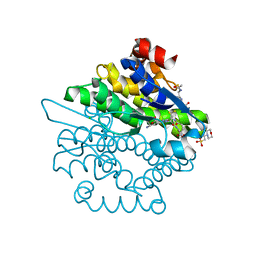 | | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUTATHIONE, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-06-17 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit
To be published
|
|
5TCC
 
 | | Complement Factor D inhibited with JH4 | | 分子名称: | (2S)-N-(6-bromopyridin-2-yl)-3-[(1H-indazol-1-yl)acetyl]-1,3-thiazolidine-2-carboxamide, Complement factor D | | 著者 | Stuckey, J.A. | | 登録日 | 2016-09-14 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.37 Å) | | 主引用文献 | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5SYM
 
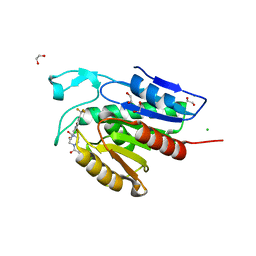 | | Cocrystal structure of the human acyl protein thioesterase 1 with an isoform-selective inhibitor, ML348 | | 分子名称: | 1,2-ETHANEDIOL, Acyl-protein thioesterase 1, CHLORIDE ION, ... | | 著者 | Stuckey, J.A, Labby, K.J, Meagher, J.L, Won, S.J, Martin, B.R. | | 登録日 | 2016-08-11 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2).
ACS Chem. Biol., 11, 2016
|
|
4LB0
 
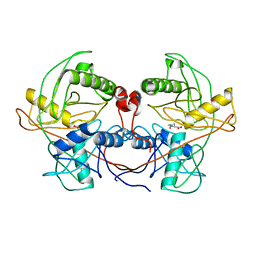 | | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline | | 分子名称: | 4-HYDROXYPROLINE, ACETATE ION, Uncharacterized protein | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-06-20 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline
To be Published
|
|
4LC6
 
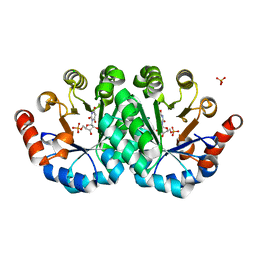 | | Crystal structure of the mutant H128Q of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2013-06-21 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Crystal structure of the mutant H128Q of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4L8G
 
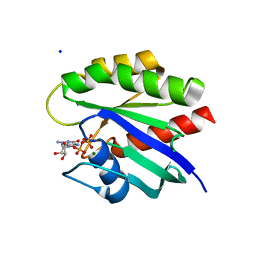 | | Crystal Structure of K-Ras G12C, GDP-bound | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | 登録日 | 2013-06-17 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.521 Å) | | 主引用文献 | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4KZE
 
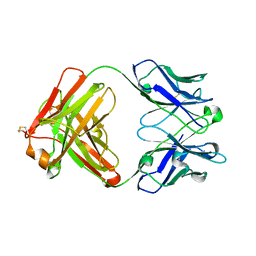 | | Crystal structure of an RNA aptamer in complex with Fab | | 分子名称: | BL3-6 Fab antibody, heavy chain, light chain, ... | | 著者 | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | 登録日 | 2013-05-29 | | 公開日 | 2014-06-18 | | 最終更新日 | 2014-07-30 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
