4I9Z
 
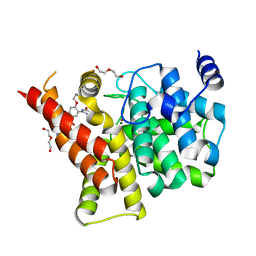 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | 分子名称: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Ren, J, Chen, T, Xu, Y. | | 登録日 | 2012-12-05 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5DDV
 
 | |
3OHI
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | 分子名称: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | 著者 | Herzberg, O, Galkin, A. | | 登録日 | 2010-08-17 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | 分子名称: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2019-02-01 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | 分子名称: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2019-01-04 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
1MB6
 
 | |
6M12
 
 | | Crystal Structure of Rnase L in complex with SU11652 | | 分子名称: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2020-02-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M13
 
 | | Crystal structure of Rnase L in complex with Toceranib | | 分子名称: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2020-02-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
7Q6T
 
 | | Crystal structure of the bromodomain of ATAD2 with AZ13824374 | | 分子名称: | (1R,9S,12R)-13-[[8-[[1-(2-fluoranyl-2-methyl-propyl)piperidin-4-yl]amino]-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-12-propan-2-yl-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | 著者 | Patel, S.J, Winter-Holt, J.J. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6W
 
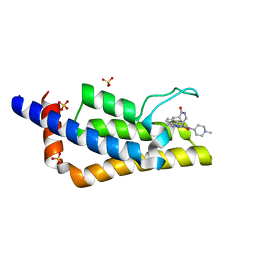 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridazine (cpd 22) | | 分子名称: | (1R,9S)-13-[[3-methyl-8-[(1-methylpiperidin-4-yl)amino]-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | 著者 | Patel, S.J, Winter-Holt, J.J. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6U
 
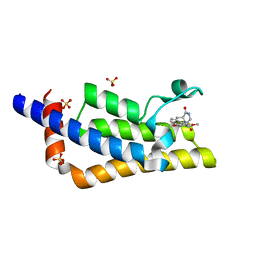 | | Crystal structure of the bromodomain of ATAD2 with phenol HTS hit (cpd 6) | | 分子名称: | (1R,9S)-13-(3,5-dimethoxy-4-oxidanyl-phenyl)carbonyl-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | 著者 | Patel, S.J, Winter-Holt, J.J. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6V
 
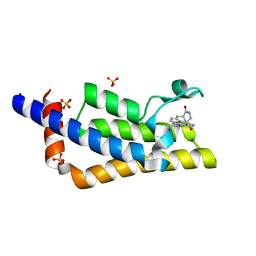 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridine (cpd 14) | | 分子名称: | (1R,9S)-13-[(8-azanyl-3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | 著者 | Patel, S.J, Winter-Holt, J.J. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
6LN0
 
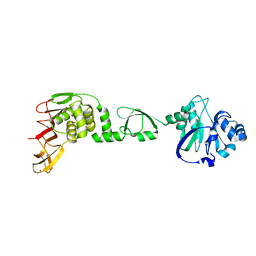 | |
7QPB
 
 | | Catalytic C-lobe of the HECT-type ubiquitin ligase E6AP in complex with a hybrid foldamer-peptide macrocycle | | 分子名称: | Isoform I of Ubiquitin-protein ligase E3A, hybrid foldamer-peptide macrocycle | | 著者 | Dengler, S, Howard, R.T, Morozov, V, Tsiamantas, C, Douat, C, Suga, H, Huc, I. | | 登録日 | 2022-01-03 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.342 Å) | | 主引用文献 | Display Selection of a Hybrid Foldamer-Peptide Macrocycle.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7R7N
 
 | |
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | 登録日 | 2021-06-23 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7RJC
 
 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJE
 
 | | Complex III2 from Candida albicans, Inz-5 bound | | 分子名称: | 3-[2-fluoro-5-(trifluoromethyl)phenyl]-7-methyl-1-[(2-methyl-2H-tetrazol-5-yl)methyl]-1H-indazole, Cytochrome b, Cytochrome b-c1 complex subunit 2, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJD
 
 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in c position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJB
 
 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in b position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJA
 
 | |
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | 分子名称: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | 著者 | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | 分子名称: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | 著者 | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-09 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
3PJ6
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | 分子名称: | HIV protease | | 著者 | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | 登録日 | 2010-11-08 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
