6LLB
 
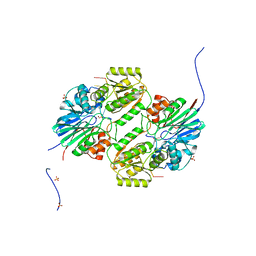 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | 分子名称: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | 著者 | Li, D.F, Hou, Y.J, Guo, L. | | 登録日 | 2019-12-22 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
7E0B
 
 | |
8FPJ
 
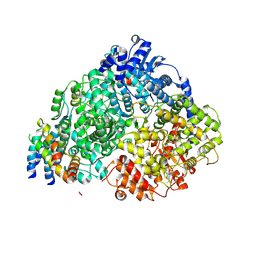 | |
8FPI
 
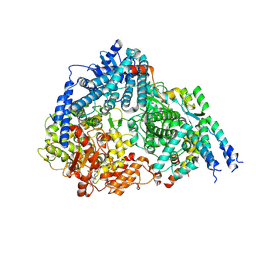 | |
6PM9
 
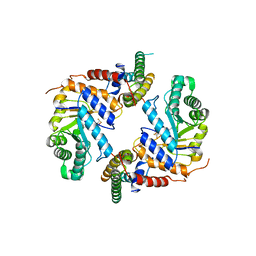 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | 分子名称: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | 著者 | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | 登録日 | 2019-07-01 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
7WZS
 
 | |
5YO5
 
 | |
5YOE
 
 | | Crystal Structure of flavodoxin with engineered disulfide bond A43C-L74C | | 分子名称: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | 著者 | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | 登録日 | 2017-10-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YM7
 
 | |
5YO3
 
 | | Crystal Structure of B562RIL with engineered disulfide bond V16C-A29C | | 分子名称: | SULFATE ION, Soluble cytochrome b562 | | 著者 | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YO6
 
 | |
5YOB
 
 | | Crystal Structure of flavodoxin without engineered disulfide bond | | 分子名称: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | 著者 | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | 登録日 | 2017-10-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.142 Å) | | 主引用文献 | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5Z6Q
 
 | |
6W4Q
 
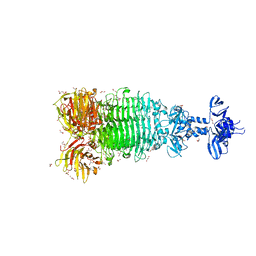 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Greenfield, J, Herzberg, O. | | 登録日 | 2020-03-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | 分子名称: | 8-amino-7-oxononanoate synthase | | 著者 | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | 登録日 | 2014-08-20 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
7RWR
 
 | | An RNA aptamer that decreases flavin redox potential | | 分子名称: | FLAVIN MONONUCLEOTIDE, RNA (38-MER) | | 著者 | Gremminger, T, Li, J, Chen, S, Heng, X. | | 登録日 | 2021-08-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An RNA aptamer that shifts the reduction potential of metabolic cofactors.
Nat.Chem.Biol., 18, 2022
|
|
5ZVL
 
 | |
3WBM
 
 | | Crystal Structure of protein-RNA complex | | 分子名称: | DNA/RNA-binding protein Alba 1, RNA (25-MER) | | 著者 | Ding, J, Wang, D.C. | | 登録日 | 2013-05-20 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea.
J.Biol.Chem., 289, 2014
|
|
8X5D
 
 | |
4QXE
 
 | |
6IUP
 
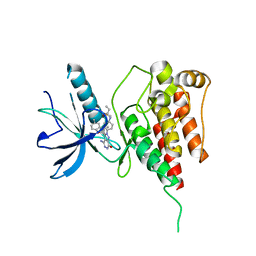 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | 分子名称: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | 著者 | Xu, Y, Liu, Q. | | 登録日 | 2018-11-29 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
7KDT
 
 | |
3E51
 
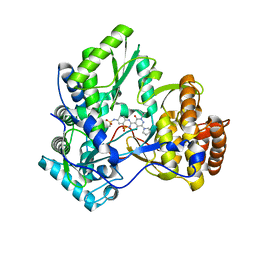 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | 分子名称: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-pyrrolidin-1-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | 著者 | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | 登録日 | 2008-08-12 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 5: Exploration of pyridazinones containing 6-amino-substituents.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4TY1
 
 | |
4U93
 
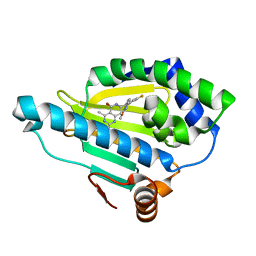 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | 分子名称: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | 著者 | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | 登録日 | 2014-08-05 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
