8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | 分子名称: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | 著者 | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | 登録日 | 2023-07-03 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.33 Å) | | 主引用文献 | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
7XYB
 
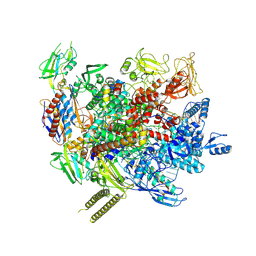 | | The cryo-EM structure of an AlpA-loaded complex | | 分子名称: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Wen, A, Feng, Y. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XYA
 
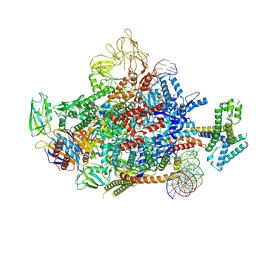 | | The cryo-EM structure of an AlpA-loading complex | | 分子名称: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Wen, A, Feng, Y. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XM0
 
 | |
7MWH
 
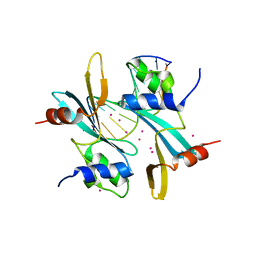 | | Crystal structure of BAZ2A with DNA | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | 著者 | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2021-05-17 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
8H3I
 
 | |
8H3J
 
 | |
7S1N
 
 | |
6OAG
 
 | |
6N7Y
 
 | |
6N83
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | 分子名称: | CHLORIDE ION, Farnesyl pyrophosphate synthase, PHOSPHATE ION, ... | | 著者 | Park, J, Schilling, M.A, Berghuis, A.M. | | 登録日 | 2018-11-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
6N7Z
 
 | |
6N82
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Farnesyl pyrophosphate synthase, ... | | 著者 | Park, J, Schilling, M.A, Berghuis, A.M. | | 登録日 | 2018-11-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
6OAH
 
 | |
4KFV
 
 | |
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | 分子名称: | Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | 登録日 | 2019-04-21 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.233 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.293 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-20 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.183 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
4KFW
 
 | |
8GB6
 
 | |
8GB7
 
 | |
8GB8
 
 | |
8GB5
 
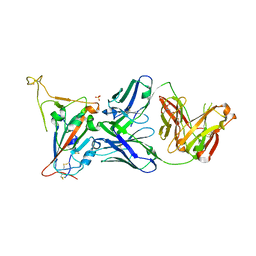 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | 分子名称: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2023-02-24 | | 公開日 | 2023-05-24 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
7Y67
 
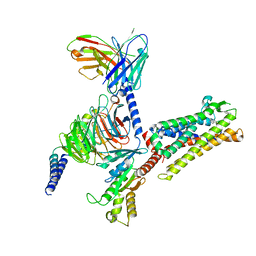 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | 分子名称: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
