3UDW
 
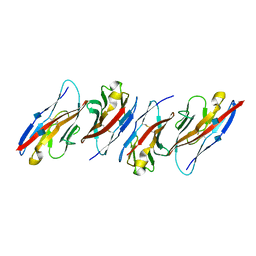 | | Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T cell immunoreceptor with Ig and ITIM domains | | 著者 | Rouge, L, Stengel, K.F, Yin, J.P, Bazan, F.J, Wiesmann, C. | | 登録日 | 2011-10-28 | | 公開日 | 2012-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.903 Å) | | 主引用文献 | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | 分子名称: | NEQ131 | | 著者 | Zhang, J, Gan, J. | | 登録日 | 2016-05-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
3LGD
 
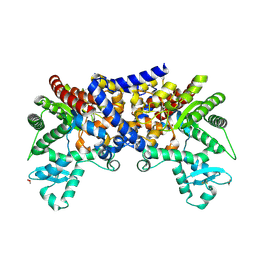 | |
3LGG
 
 | | Crystal structure of human adenosine deaminase growth factor, adenosine deaminase type 2 (ADA2) complexed with transition state analogue, coformycin | | 分子名称: | (8R)-3-beta-D-ribofuranosyl-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase CECR1, ... | | 著者 | Zavialov, A.V. | | 登録日 | 2010-01-20 | | 公開日 | 2010-02-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the growth factor activity of human adenosine deaminase ADA2.
J.Biol.Chem., 285, 2010
|
|
5IJ8
 
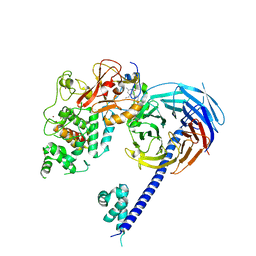 | | Structure of the primary oncogenic mutant Y641N Hs/AcPRC2 in complex with a pyridone inhibitor | | 分子名称: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | 著者 | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | 登録日 | 2016-03-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
5YAD
 
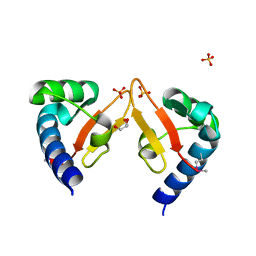 | |
5YAA
 
 | |
5JRC
 
 | |
5YBI
 
 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | 著者 | Mu, Z.X, Gao, X.P, Cui, S. | | 登録日 | 2017-09-05 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.268 Å) | | 主引用文献 | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | 分子名称: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.254 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | 著者 | Wang, H, Liu, X, Sun, L, Yang, H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | 分子名称: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
5JRE
 
 | |
7XCZ
 
 | |
7XDA
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | 著者 | Liu, Z, Lui, S, Gao, Y. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | 著者 | Liu, Z, Liu, S, Gao, Y.Z. | | 登録日 | 2022-03-26 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | 著者 | Liu, Z, Liu, S, Yuanzhu, G. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | 分子名称: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | 著者 | Wang, H, Li, D, Sun, L, Yang, H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | 分子名称: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | 著者 | Wang, H, Shao, M, Sun, L, Yang, H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | 分子名称: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Wang, H, Li, D, Sun, L, Yang, H. | | 登録日 | 2022-11-07 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | 分子名称: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | 著者 | Wang, H, Shao, M, Sun, L, Yang, H. | | 登録日 | 2022-11-12 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | 分子名称: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | 著者 | Wang, H, Li, D, Sun, L, Yang, H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
2MB9
 
 | | Human Bcl10 CARD | | 分子名称: | B-cell lymphoma/leukemia 10 | | 著者 | Zheng, C, Bracken, C, Wu, H. | | 登録日 | 2013-07-26 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
4HJI
 
 | |
