3IKI
 
 | | 5-SMe-dU containing DNA octamer | | 分子名称: | 5'-D(*GP*(UMS)P*GP*(US2)P*AP*CP*AP*C)-3', MAGNESIUM ION | | 著者 | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | 登録日 | 2009-08-05 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
3IT9
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in apo state | | 分子名称: | D-alanyl-D-alanine carboxypeptidase dacC, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | 登録日 | 2009-08-27 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3IJN
 
 | |
3HG8
 
 | |
3HGD
 
 | |
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | 分子名称: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | 著者 | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | 登録日 | 2009-08-27 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3J0F
 
 | | Sindbis virion | | 分子名称: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | 著者 | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | 登録日 | 2011-07-08 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
8IQU
 
 | | Structure of MtbFadD23 with PhU-AMS | | 分子名称: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Fatty-acid-CoA ligase FadD23 | | 著者 | Yan, M.R, Zhang, W. | | 登録日 | 2023-03-17 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural basis for the development of potential inhibitors targeting FadD23 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8JJM
 
 | |
8FZC
 
 | |
7DW5
 
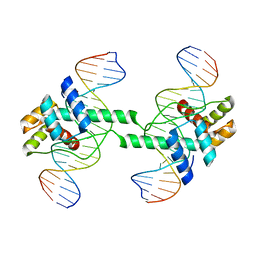 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | 分子名称: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | 著者 | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | 登録日 | 2021-01-15 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
3TFZ
 
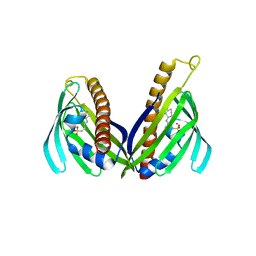 | | Crystal structure of Zhui aromatase/cyclase from Streptomcyes sp. R1128 | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclase, POTASSIUM ION | | 著者 | Ames, B.D, Lee, M.Y, Moody, C, Zhang, W, Tang, Y, Wong, S.K, Tsai, S.C. | | 登録日 | 2011-08-16 | | 公開日 | 2011-09-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural and Biochemical Characterization of ZhuI Aromatase/Cyclase from the R1128 Polyketide Pathway.
Biochemistry, 50, 2011
|
|
8OG2
 
 | | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A | | 分子名称: | COENZYME A, SODIUM ION, SULFATE ION, ... | | 著者 | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | 登録日 | 2023-03-17 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
7U0N
 
 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | 登録日 | 2022-02-18 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
7WA9
 
 | |
7ESH
 
 | | Crystal structure of amylosucrase from Calidithermus timidus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | 著者 | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | 登録日 | 2021-05-10 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
7DEA
 
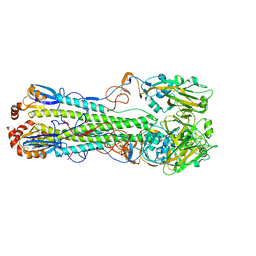 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | 登録日 | 2020-11-03 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
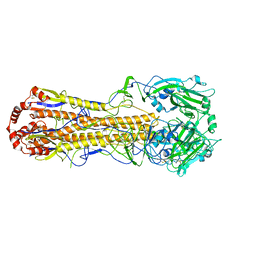 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | 登録日 | 2020-11-03 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7C0N
 
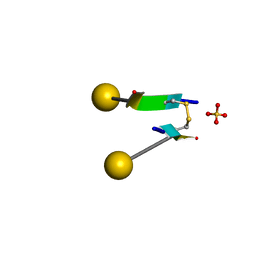 | | Crystal structure of a self-assembling galactosylated peptide homodimer | | 分子名称: | SULFATE ION, Self-assembling galactosylated tyrosine-rich peptide, beta-D-galactopyranose | | 著者 | He, C, Wu, S, Chi, C, Zhang, W, Ma, M, Lai, L, Dong, S. | | 登録日 | 2020-05-01 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-10-21 | | 実験手法 | X-RAY DIFFRACTION (1.552 Å) | | 主引用文献 | Glycopeptide Self-Assembly Modulated by Glycan Stereochemistry through Glycan-Aromatic Interactions.
J.Am.Chem.Soc., 142, 2020
|
|
5V69
 
 | |
5V5G
 
 | |
5V5I
 
 | |
5NS9
 
 | |
6FE8
 
 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | 著者 | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | 登録日 | 2017-12-30 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
3B6T
 
 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | 著者 | Cho, Y, Lolis, E, Howe, J.R. | | 登録日 | 2007-10-29 | | 公開日 | 2008-02-05 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
