6C91
 
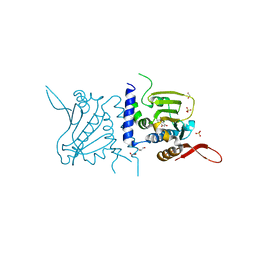 | | Structure of GRP94 with a resorcinylic inhibitor. | | 分子名称: | 1,2-ETHANEDIOL, 5-[2-(1-benzyl-1H-imidazol-2-yl)ethyl]-4,6-dichlorobenzene-1,3-diol, Endoplasmin, ... | | 著者 | Que, N.L.S, Gewirth, D.T. | | 登録日 | 2018-01-25 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.895 Å) | | 主引用文献 | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8KAE
 
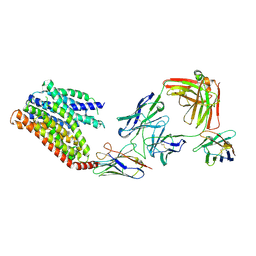 | | 16d-bound human SPNS2 | | 分子名称: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | 著者 | He, Y, Duan, Y. | | 登録日 | 2023-08-03 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
8K3K
 
 | |
6LYC
 
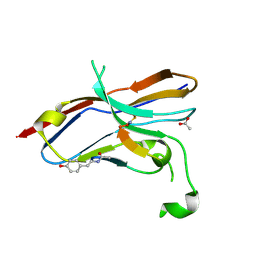 | | Crystal structure of the NOD SIRPa complex with D4-2 | | 分子名称: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | 著者 | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | 登録日 | 2020-02-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
5H3R
 
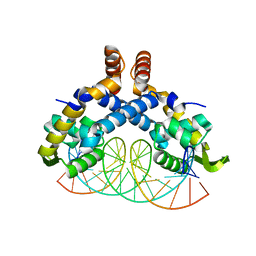 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | 著者 | Zhu, R, Lou, H, Hao, Z. | | 登録日 | 2016-10-26 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7VAH
 
 | |
8WGW
 
 | | Local refinement of RBD-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
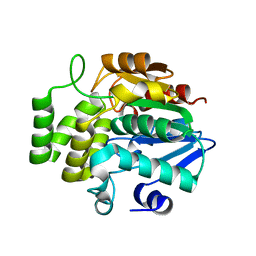
7F5W
 
 | |
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
5JLY
 
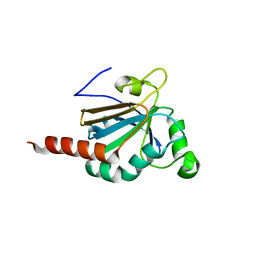 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | 分子名称: | Thioredoxin peroxidase-1 | | 著者 | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | 登録日 | 2016-04-27 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.051 Å) | | 主引用文献 | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
7W9N
 
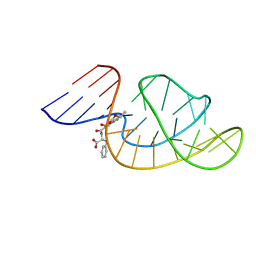 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | 著者 | Xu, G.H, Li, C.G. | | 登録日 | 2021-12-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
2JG4
 
 | | Substrate-free IDE structure in its closed conformation | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | 著者 | Malito, E, Tang, W.J. | | 登録日 | 2007-02-07 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.396 Å) | | 主引用文献 | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
2JBU
 
 | | Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CO-PURIFIED PEPTIDE, INSULIN-DEGRADING ENZYME | | 著者 | Im, H, Shen, Y, Tang, W.J. | | 登録日 | 2006-12-11 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
4J01
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | 分子名称: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.246 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J00
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | 分子名称: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.004 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | 分子名称: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Zhang, J, Huang, Z, Song, K. | | 登録日 | 2017-07-25 | | 公開日 | 2018-11-07 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
7E5X
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
