7DGE
 
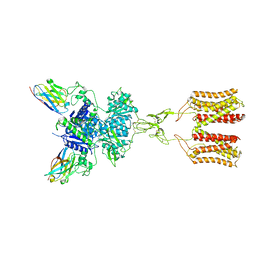 | | intermediate state of class C GPCR | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 1, nanobody | | 著者 | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | 登録日 | 2020-11-11 | | 公開日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
4HFQ
 
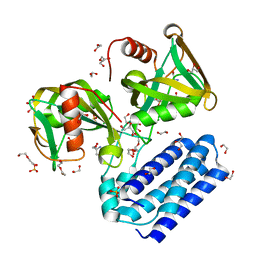 | | Crystal structure of UDP-X diphosphatase | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | 登録日 | 2012-10-05 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | 分子名称: | ACETATE ION, Oxidoreductase, SULFATE ION | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-09 | | 公開日 | 2012-11-28 | | 最終更新日 | 2016-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | 分子名称: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | 分子名称: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DPM
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW06 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-12-20 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.304 Å) | | 主引用文献 | Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Mabs, 13, 2021
|
|
7CE2
 
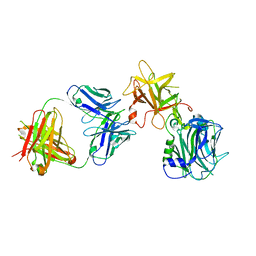 | | The Crystal structure of TeNT Hc complexed with neutralizing antibody | | 分子名称: | Tetanus toxin, neutralizing antibody heavy chain, neutralizing antibody light chain | | 著者 | Wang, X, Wang, Y, Wu, C, Yu, J, Liao, H. | | 登録日 | 2020-06-21 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural basis of tetanus toxin neutralization by native human monoclonal antibodies.
Cell Rep, 35, 2021
|
|
7CT5
 
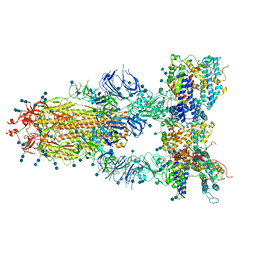 | | S protein of SARS-CoV-2 in complex bound with T-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Guo, L, Bi, W.W, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q, Dang, B.B. | | 登録日 | 2020-08-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Cell Res., 31, 2021
|
|
6JDE
 
 | |
6IWV
 
 | |
6JGY
 
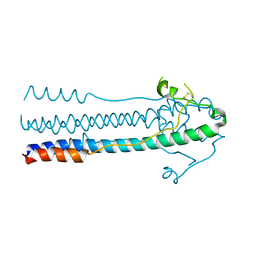 | | Crystal structure of LASV-GP2 in a post fusion conformation | | 分子名称: | Pre-glycoprotein polyprotein GP complex | | 著者 | Zhu, Y, Zhang, X, Chen, B, Ye, S, Zhang, R. | | 登録日 | 2019-02-15 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.389 Å) | | 主引用文献 | Crystal Structure of Refolding Fusion Core of Lassa Virus GP2 and Design of Lassa Virus Fusion Inhibitors.
Front Microbiol, 10, 2019
|
|
6JRK
 
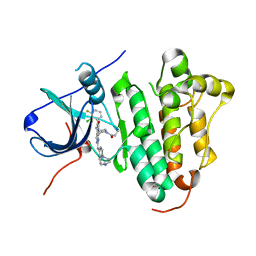 | | The structure of co-crystals of 8r-B-EGFR WT complex | | 分子名称: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | 著者 | Zhu, S.J, Yun, C.H. | | 登録日 | 2019-04-04 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.796 Å) | | 主引用文献 | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRJ
 
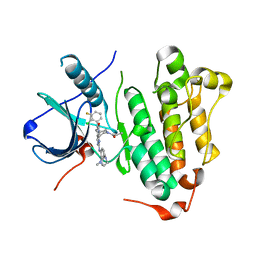 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | 分子名称: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | 著者 | Zhu, S.J, Yun, C.H. | | 登録日 | 2019-04-04 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.943 Å) | | 主引用文献 | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6A0A
 
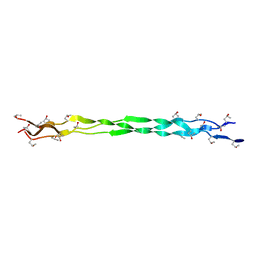 | |
5ZF2
 
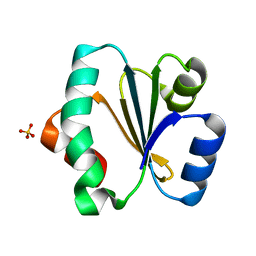 | |
6A0C
 
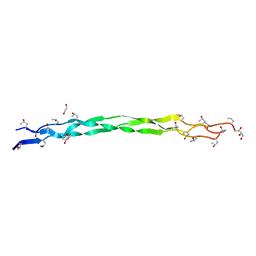 | | Structure of a triple-helix region of human collagen type III | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, collagen type III peptide | | 著者 | Yang, X, Zhu, Y, Ye, S, Zhang, R. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | 著者 | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-12-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.0012 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
8H5Z
 
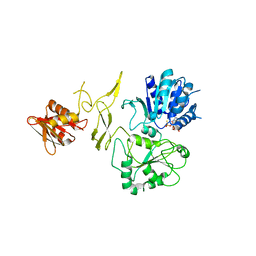 | | Crystal structure of RadD/ATP analogue complex | | 分子名称: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative DNA repair helicase RadD, ZINC ION | | 著者 | Yan, X.X, Tian, L.F. | | 登録日 | 2022-10-14 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.00002718 Å) | | 主引用文献 | Biochemical and Structural Analyses Shed Light on the Mechanisms of RadD DNA Binding and Its ATPase from Escherichia coli.
Int J Mol Sci, 24, 2023
|
|
8H5Y
 
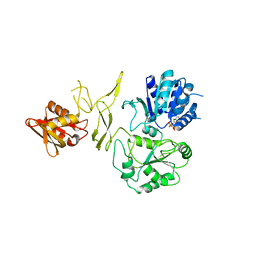 | | Crystal structure of RadD- ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | 著者 | Yan, X.X, Tian, L.F. | | 登録日 | 2022-10-14 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7001 Å) | | 主引用文献 | Biochemical and Structural Analyses Shed Light on the Mechanisms of RadD DNA Binding and Its ATPase from Escherichia coli.
Int J Mol Sci, 24, 2023
|
|
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, EPITHELIAL-CADHERIN | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
7UF8
 
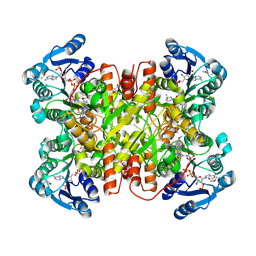 | | Structure of CtdP in complex with penicimutamide E and NADP+ | | 分子名称: | 1,2-ETHANEDIOL, CtdP, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Rivera, S, Liu, Z, Newmister, S.A, Gao, X, Sherman, D.H. | | 登録日 | 2022-03-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | An NmrA-like enzyme-catalysed redox-mediated Diels-Alder cycloaddition with anti-selectivity.
Nat.Chem., 15, 2023
|
|
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W14
 
 | |
7VYL
 
 | |
