6XQI
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | 分子名称: | ASN-PRO-LEU-GLU-PHE-LEU, Protein Vpr, UV excision repair protein RAD23 homolog A, ... | | 著者 | Calero, G.C, Wu, Y, Weiss, S.C. | | 登録日 | 2020-07-09 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
6XQJ
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | 分子名称: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | 著者 | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | 登録日 | 2020-07-09 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
7VTC
 
 | | Crystal structure of MERS main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2021-10-28 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.53865623 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLQ
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.939106 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
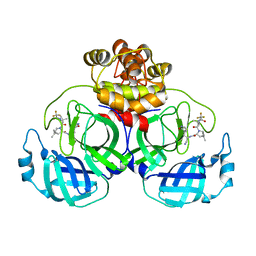 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.0227 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.50251937 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7AMN
 
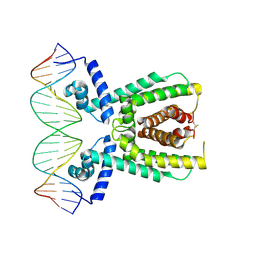 | | Structure of LuxR with DNA (repression) | | 分子名称: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | 著者 | Liu, B, Reverter, D. | | 登録日 | 2020-10-09 | | 公開日 | 2021-03-31 | | 最終更新日 | 2021-04-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7AMT
 
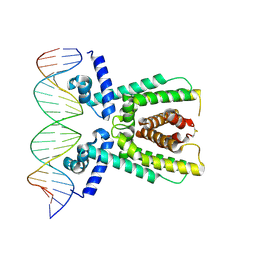 | | Structure of LuxR with DNA (activation) | | 分子名称: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | 著者 | Liu, B, Reverter, D. | | 登録日 | 2020-10-09 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
6EN8
 
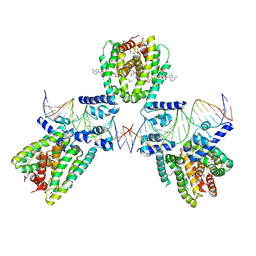 | | SaFadR in complex with dsDNA | | 分子名称: | DNA (5'-D(*CP*TP*AP*CP*TP*TP*GP*AP*TP*TP*TP*TP*TP*GP*AP*GP*TP*CP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*AP*CP*TP*CP*AP*AP*AP*AP*AP*TP*CP*AP*AP*GP*TP*AP*G)-3'), Transcriptional regulator TetR family, ... | | 著者 | Valegard, K. | | 登録日 | 2017-10-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius.
Nat Commun, 10, 2019
|
|
6EL2
 
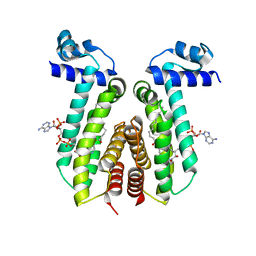 | | SaFadR_lauroyl_CoA complex | | 分子名称: | DODECYL-COA, Transcriptional regulator TetR family | | 著者 | Valegard, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius.
Nat Commun, 10, 2019
|
|
7VFB
 
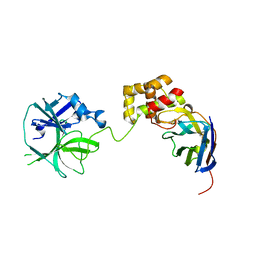 | |
7VFA
 
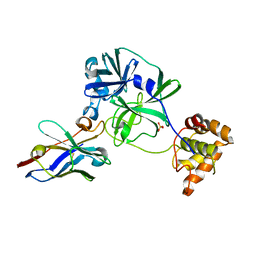 | |
6MZP
 
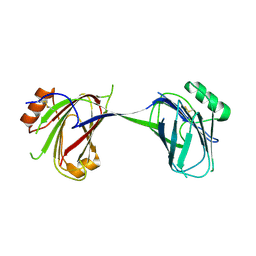 | |
6MZN
 
 | |
7RJF
 
 | | MOPD-1 mutant-L47W | | 分子名称: | MALONATE ION, ZINC ION, [L47W]MOPD-1 | | 著者 | Huawu, Y, Conan, K.W, Gordon, J.K, Brett, M.C, Yen-Hua, H, David, J.C. | | 登録日 | 2021-07-20 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rational Design of Potent Peptide Inhibitors of the PD-1:PD-L1 Interaction for Cancer Immunotherapy.
J.Am.Chem.Soc., 143, 2021
|
|
7VSW
 
 | |
7VSU
 
 | |
6IEV
 
 | | Crystal structure of a designed protein | | 分子名称: | Designed protein | | 著者 | Han, M, Liao, S, Chen, Q, Liu, H. | | 登録日 | 2018-09-17 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
3R9X
 
 | | Crystal structure of Era in complex with MgGDPNP, nucleotides 1506-1542 of 16S ribosomal RNA, and KsgA | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, GTPase Era, ... | | 著者 | Tu, C, Ji, X. | | 登録日 | 2011-03-26 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R9W
 
 | |
8K6Q
 
 | | Crystal structure of HOIL-1L LTM domain | | 分子名称: | RanBP-type and C3HC4-type zinc finger-containing protein 1 | | 著者 | Yan, Z, Pan, L.F. | | 登録日 | 2023-07-25 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Mechanistic insights into the homo-dimerization of HOIL-1L and SHARPIN.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8K6P
 
 | |
8UPV
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | 分子名称: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | 著者 | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | 著者 | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | 分子名称: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | 著者 | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
