3VBR
 
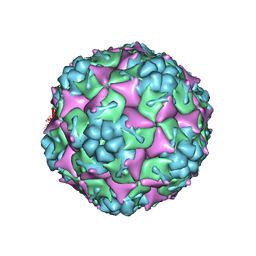 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | 分子名称: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | 著者 | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | 登録日 | 2012-01-02 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBF
 
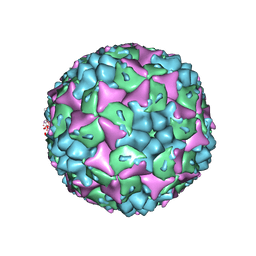 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | 分子名称: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | 著者 | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | 登録日 | 2012-01-02 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBO
 
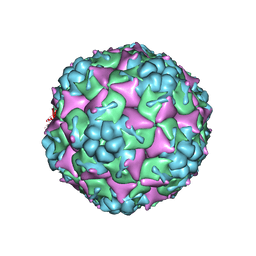 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (cryo at 100K) | | 分子名称: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | 著者 | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | 登録日 | 2012-01-02 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2LCW
 
 | |
4QPG
 
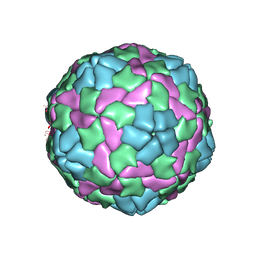 | | Crystal structure of empty hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4KQQ
 
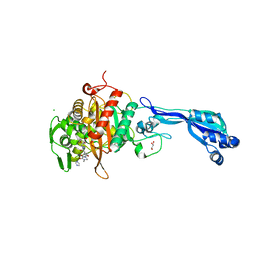 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | 分子名称: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | 登録日 | 2013-05-15 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | 分子名称: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | 著者 | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | 登録日 | 2013-05-15 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | 分子名称: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | 登録日 | 2013-05-15 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
5HNS
 
 | | Structure of glycosylated NPC1 luminal domain C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | 著者 | Zhao, Y, Ren, J, Harlos, K, Stuart, D.I. | | 登録日 | 2016-01-18 | | 公開日 | 2016-02-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions.
Febs Lett., 590, 2016
|
|
4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | 登録日 | 2014-04-14 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4Q4B
 
 | | Crystal structure of LIMP-2 (space group C2221) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | 著者 | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | 登録日 | 2014-04-14 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4I2X
 
 | | Crystal structure of Signal Regulatory Protein gamma (SIRP-gamma) in complex with FabOX117 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FabOX117 heavy chain, ... | | 著者 | Nettleship, J.E, Ren, J, Stuart, D.I, Owens, R.J, Oxford Protein Production Facility (OPPF) | | 登録日 | 2012-11-23 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal structure of signal regulatory protein gamma (SIRP gamma) in complex with an antibody Fab fragment.
Bmc Struct.Biol., 13, 2013
|
|
4ILE
 
 | |
4IV1
 
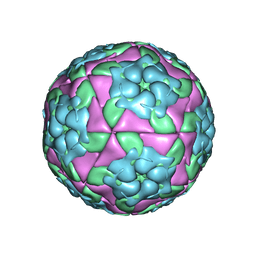 | | Crystal structure of recombinant foot-and-mouth-disease virus A22 empty capsid | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | 登録日 | 2013-01-22 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
4IV3
 
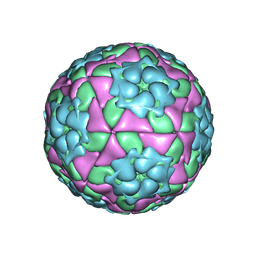 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | 登録日 | 2013-01-22 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
3OC2
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | 分子名称: | CHLORIDE ION, Penicillin-binding protein 3 | | 著者 | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | 登録日 | 2010-08-09 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.968 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCN
 
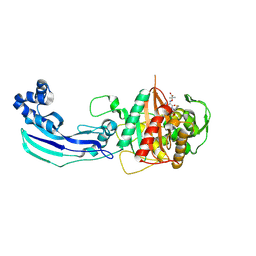 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with ceftazidime | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, penicillin-binding protein 3 | | 著者 | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | 登録日 | 2010-08-10 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCL
 
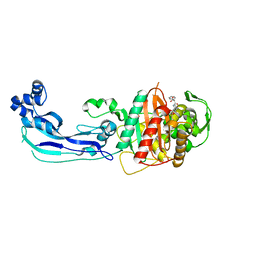 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with carbenicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | 登録日 | 2010-08-10 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
4AB5
 
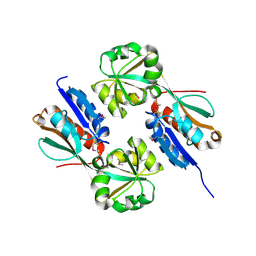 | | Regulatory domain structure of NMB2055 (MetR) a LysR family regulator from N. meningitidis | | 分子名称: | TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | 著者 | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | 登録日 | 2011-12-07 | | 公開日 | 2012-07-11 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4AB6
 
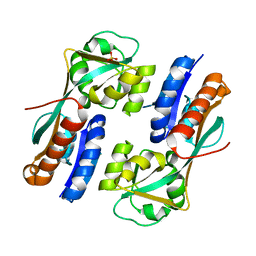 | | Regulatory domain structure of NMB2055 (MetR), C103S C106S mutant, a LysR family regulator from N. meningitidis | | 分子名称: | SULFATE ION, TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | 著者 | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | 登録日 | 2011-12-07 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4WYU
 
 | |
4WYT
 
 | |
5ZBS
 
 | |
5ZBR
 
 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | 分子名称: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Ren, J.Q, Wang, S, Feng, W. | | 登録日 | 2018-02-12 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
