3EUH
 
 | | Crystal Structure of the MukE-MukF Complex | | 分子名称: | Chromosome partition protein mukF, GLYCINE, MukE | | 著者 | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | 登録日 | 2008-10-10 | | 公開日 | 2009-01-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
4AWP
 
 | |
3R4Y
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | 分子名称: | Glycosyl hydrolase family 32, N terminal | | 著者 | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | 登録日 | 2011-03-18 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Caldwell, J.P, Strickland, C. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R95
 
 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R92
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R8Y
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Caldwell, J.P, Strickland, C. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R93
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-1-methyl-5-oxo-4-(((1S,3R)-3-(3-phenylureido)cyclohexyl)methyl)imidazolidin-2-iminium | | 分子名称: | 1-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]-3-phenylurea, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | 分子名称: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | 著者 | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | 登録日 | 2011-03-18 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
2QBR
 
 | | Crystal structure of ptp1b-inhibitor complex | | 分子名称: | 5-[3-(BENZYLAMINO)PHENYL]-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Xu, W. | | 登録日 | 2007-06-18 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBQ
 
 | | Crystal structure of ptp1b-inhibitor complex | | 分子名称: | 4-BROMO-3-(CARBOXYMETHOXY)-5-{3-[(3,3,5,5-TETRAMETHYLCYCLOHEXYL)AMINO]PHENYL}THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Xu, W. | | 登録日 | 2007-06-18 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBP
 
 | | Crystal structure of ptp1b-inhibitor complex | | 分子名称: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Xu, W. | | 登録日 | 2007-06-18 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
5X90
 
 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | 分子名称: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | 著者 | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | 登録日 | 2017-03-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
5C27
 
 | | Crystal structure of SYK in complex with compound 2 | | 分子名称: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | 著者 | Han, S, Chang, J. | | 登録日 | 2015-06-15 | | 公開日 | 2015-10-07 | | 最終更新日 | 2016-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
5C26
 
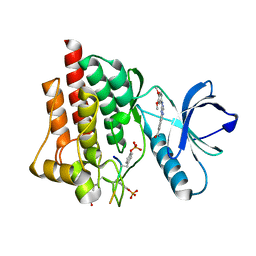 | | Crystal structure of SYK in complex with compound 1 | | 分子名称: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | 著者 | Han, S, Chang, J. | | 登録日 | 2015-06-15 | | 公開日 | 2015-10-07 | | 最終更新日 | 2016-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
2KAL
 
 | | NMR structure of fully methylated GATC site | | 分子名称: | 5'-D(*DCP*DGP*DCP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DCP*DGP*DC)-3', 5'-D(*DGP*DCP*DGP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DGP*DCP*DG)-3' | | 著者 | Bang, J, Bae, S, Park, C, Lee, J, Choi, B. | | 登録日 | 2008-11-09 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites.
J.Am.Chem.Soc., 130, 2008
|
|
7R40
 
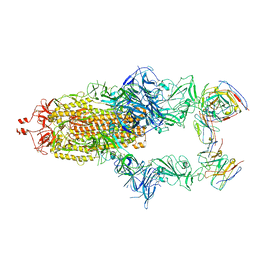 | |
3P2F
 
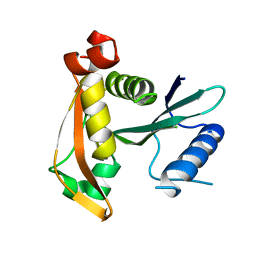 | |
3P2H
 
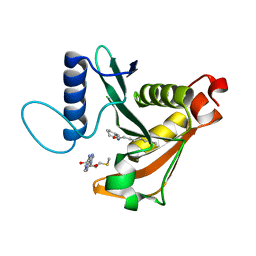 | |
3OUM
 
 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | 分子名称: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | 著者 | Kim, M.I, Rhee, S. | | 登録日 | 2010-09-15 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
3OUL
 
 | |
7V6S
 
 | |
7V6U
 
 | | Crystal structure of bacterial peptidase | | 分子名称: | 1,2-ETHANEDIOL, D-MALATE, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | 著者 | Kim, Y, Lee, W.C. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | The crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | 著者 | Kim, Y, Lee, W.C. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.495 Å) | | 主引用文献 | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
5XV9
 
 | |
