1I6N
 
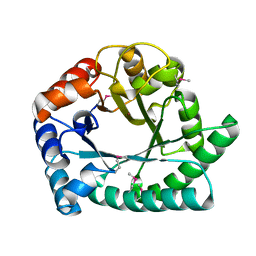 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | 分子名称: | IOLI PROTEIN, ZINC ION | | 著者 | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-03-02 | | 公開日 | 2002-03-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1DNT
 
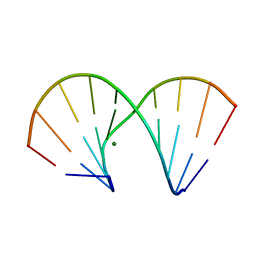 | | RNA/DNA DODECAMER R(GC)D(GTATACGC) WITH MAGNESIUM BINDING SITES | | 分子名称: | DNA/RNA (5'-R(*GP*CP)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | 著者 | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 1999-12-16 | | 公開日 | 2000-04-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1DNX
 
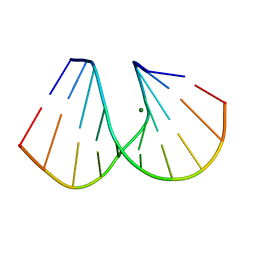 | | RNA/DNA DODECAMER R(G)D(CGTATACGC) WITH MAGNESIUM BINDING SITES | | 分子名称: | DNA/RNA (5'-R(*GP)-D(*CP*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | 著者 | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 1999-12-16 | | 公開日 | 2000-04-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
6Q2B
 
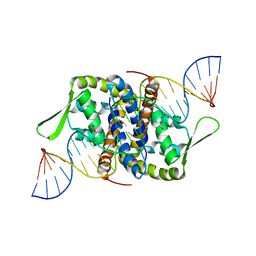 | | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA | | 分子名称: | ACETIC ACID, DNA (26-MER), MarR family transcriptional regulator | | 著者 | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA.
To Be Published
|
|
6PXA
 
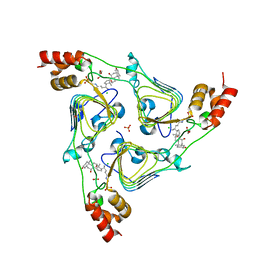 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | 分子名称: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | 著者 | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-25 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid
To Be Published
|
|
6PUB
 
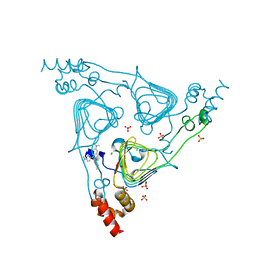 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | 分子名称: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | 著者 | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-18 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
1G60
 
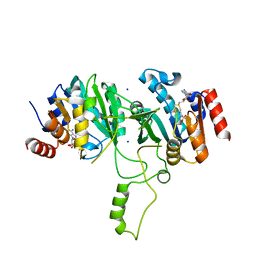 | | Crystal Structure of Methyltransferase MboIIa (Moraxella bovis) | | 分子名称: | Adenine-specific Methyltransferase MboIIA, S-ADENOSYLMETHIONINE, SODIUM ION | | 著者 | Osipiuk, J, Walsh, M.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2000-11-02 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal structure of MboIIA methyltransferase.
Nucleic Acids Res., 31, 2003
|
|
1EXZ
 
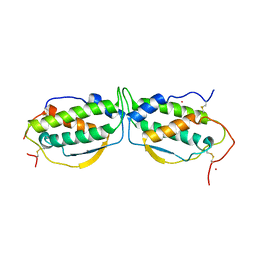 | | STRUCTURE OF STEM CELL FACTOR | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, SAMARIUM (III) ION, ... | | 著者 | Zhang, Z, Zhang, R, Joachimiak, A, Schlessinger, J, Kong, X. | | 登録日 | 2000-05-05 | | 公開日 | 2000-07-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EG4
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | 分子名称: | BETA-DYSTROGLYCAN, DYSTROPHIN | | 著者 | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | 登録日 | 2000-02-11 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EG2
 
 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | 著者 | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2000-02-11 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
1DCR
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | 分子名称: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | 著者 | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 1999-11-05 | | 公開日 | 1999-11-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1D9R
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | 分子名称: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', COBALT HEXAMMINE(III) | | 著者 | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 1999-10-29 | | 公開日 | 1999-11-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1JV2
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | 登録日 | 2001-08-28 | | 公開日 | 2001-10-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
1J4J
 
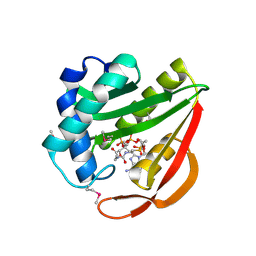 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | 分子名称: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | 著者 | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | 登録日 | 2001-10-02 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
1EG3
 
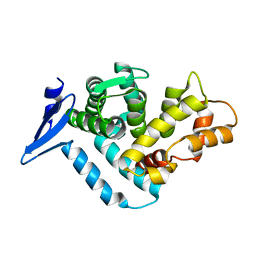 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | 分子名称: | DYSTROPHIN | | 著者 | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | 登録日 | 2000-02-11 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1CS7
 
 | | SYNTHETIC DNA HAIRPIN WITH STILBENEDIETHER LINKER | | 分子名称: | 5'-D(GP*(BRU)P*TP*TP*TP*GP*(S02)*CP*AP*AP*AP*AP*C)-3', STRONTIUM ION | | 著者 | Lewis, F.D, Liu, X, Wu, Y, Miller, S.E, Wasielewski, M.R, Letsinger, R.L, Sanishvili, R, Joachimiak, A, Tereshko, V, Egli, M. | | 登録日 | 1999-08-17 | | 公開日 | 2001-10-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and Photoinduced Electron Transfer in Exceptionally Stable Synthetic DNA Hairpins with Stilbenediether Linkers
J.Am.Chem.Soc., 121, 1999
|
|
1D8X
 
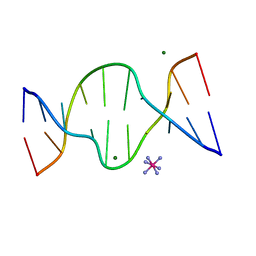 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G A BASE PAIRS | | 分子名称: | 5'-D(*CP*CP*GP*AP*AP*TP*GP*AP*GP*G)-3', COBALT HEXAMMINE(III), MAGNESIUM ION | | 著者 | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 1999-10-26 | | 公開日 | 1999-11-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
3TY7
 
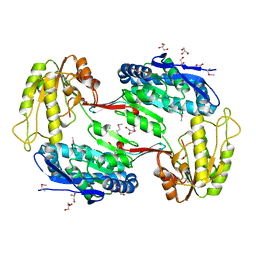 | | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kim, Y, Joachimiak, G, Jedrzejczak, R, Rubin, E, Ioerger, T, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2011-09-23 | | 公開日 | 2011-10-19 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus
To be Published
|
|
6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | 分子名称: | Lysophospholipase L1 | | 著者 | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | 分子名称: | Lysophospholipase L1, SODIUM ION | | 著者 | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
6XKH
 
 | | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine) | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, ACETATE ION, ... | | 著者 | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevsky, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine)
To Be Published
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WTC
 
 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-02 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6WKP
 
 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6XS4
 
 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | 著者 | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
