2BW1
 
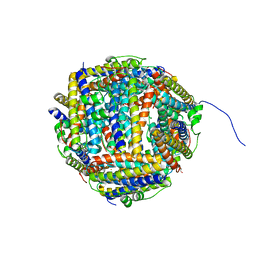 | | Iron-bound crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis. | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DPS-LIKE PEROXIDE RESISTANCE PROTEIN, ... | | 著者 | Kauko, A, Pulliainen, A, Haataja, S, Finne, J, Papageorgiou, A.C. | | 登録日 | 2005-07-07 | | 公開日 | 2006-09-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core.
J. Mol. Biol., 364, 2006
|
|
2BMC
 
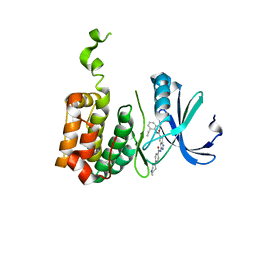 | | Aurora-2 T287D T288D complexed with PHA-680632 | | 分子名称: | (3E)-N-(2,6-DIETHYLPHENYL)-3-{[4-(4-METHYLPIPERAZIN-1-YL)BENZOYL]IMINO}PYRROLO[3,4-C]PYRAZOLE-5(3H)-CARBOXAMIDE, SERINE THREONINE-PROTEIN KINASE 6 | | 著者 | Cameron, A.D, Izzo, G, Sagliano, A, Rusconi, L, Storici, P, Fancelli, D, Berta, D, Bindi, S, Catana, C, Forte, B, Giordano, P, Mantegani, S, Meroni, M, Moll, J, Pittala, V, Severino, D, Tonani, R, Varasi, M, Vulpetti, A, Vianello, P. | | 登録日 | 2005-03-11 | | 公開日 | 2005-03-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Potent and Selective Aurora Inhibitors Identified by the Expansion of a Novel Scaffold for Protein Kinase Inhibition.
J.Med.Chem., 48, 2005
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P, Meents, A. | | 登録日 | 2020-10-21 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7SKY
 
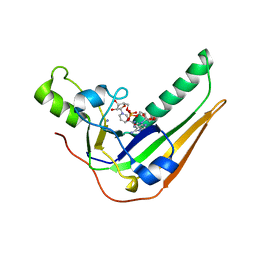 | | Pertussis toxin S1 bound to NAD+ | | 分子名称: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | 著者 | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | 登録日 | 2021-10-21 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.37000132 Å) | | 主引用文献 | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKK
 
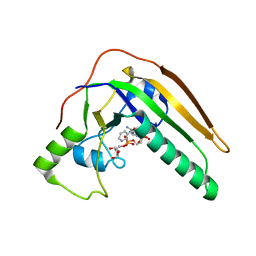 | | pertussis toxin in complex with ADPR and Nicotinamide | | 分子名称: | NICOTINAMIDE, Pertussis toxin subunit 1, SULFATE ION, ... | | 著者 | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | 登録日 | 2021-10-21 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65000772 Å) | | 主引用文献 | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8B4N
 
 | |
7ZVP
 
 | |
7ZZN
 
 | |
8A0I
 
 | |
8A08
 
 | |
8A0R
 
 | |
8A0Q
 
 | |
8A0O
 
 | |
8A0P
 
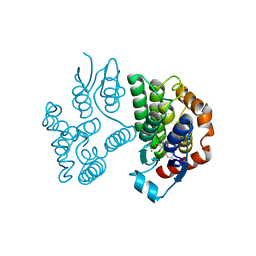 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | 分子名称: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | 著者 | Didierjean, C, Favier, F. | | 登録日 | 2022-05-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.686 Å) | | 主引用文献 | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
7ZS3
 
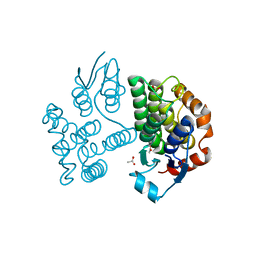 | |
7OND
 
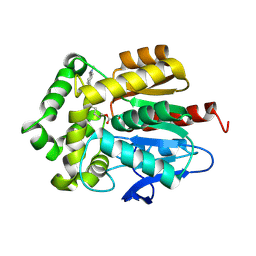 | |
7OO4
 
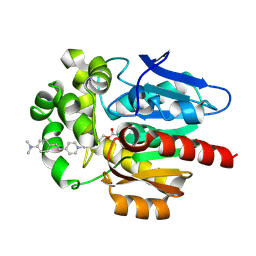 | |
8AK4
 
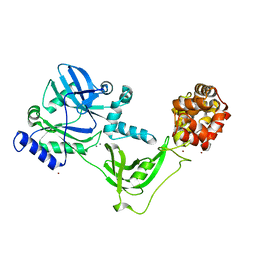 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | 分子名称: | DNA ligase, MANGANESE (II) ION, ZINC ION | | 著者 | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | 登録日 | 2022-07-29 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
4K8B
 
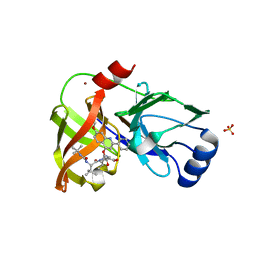 | | Crystal structure of HCV NS3/4A protease complexed with inhibitor | | 分子名称: | N-(tert-butylcarbamoyl)-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-phenylquinolin-4-yl)oxy]-L-prolinamide, NS3 protease, Nonstructural protein, ... | | 著者 | Nar, H. | | 登録日 | 2013-04-18 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Ligand bioactive conformation plays a critical role in the design of drugs that target the hepatitis C virus NS3 protease.
J.Med.Chem., 57, 2014
|
|
5T7L
 
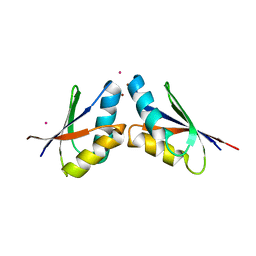 | | Pt(II)-mediated copper-dependent interactions between ATOX1 and MNK1 | | 分子名称: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1, ... | | 著者 | Caliandro, R, Mirabelli, V, Caliandro, R, Rosato, A, Lasorsa, A, Galliani, A, Arnesano, F, Natile, G. | | 登録日 | 2016-09-05 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Mechanistic and Structural Basis for Inhibition of Copper Trafficking by Platinum Anticancer Drugs.
J.Am.Chem.Soc., 141, 2019
|
|
7ZQK
 
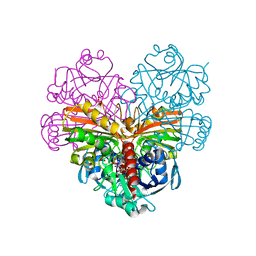 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ3
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
8VW5
 
 | | Crystal structure of Cbl-b TKB bound to compound 2 | | 分子名称: | CALCIUM ION, E3 ubiquitin-protein ligase CBL-B, MAGNESIUM ION, ... | | 著者 | Yu, C, Murray, J, Hsu, P.L. | | 登録日 | 2024-01-31 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
8VW4
 
 | | Crystal structure of Cbl-b TKB bound to compound 26 | | 分子名称: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | 著者 | Yu, C, Murray, J, Hsu, P.L. | | 登録日 | 2024-01-31 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
