4S0J
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | 分子名称: | Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-31 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S03
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79I, and F123A mutant | | 分子名称: | Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-30 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0L
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79V, and F123V mutant | | 分子名称: | Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-31 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4RRO
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4S0K
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79V, and 123A mutant | | 分子名称: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-31 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4RRS
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4R,4a'R,10a'S)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2014-12-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
3M20
 
 | | Crystal structure of DmpI from Archaeoglobus fulgidus determined to 2.37 Angstroms resolution | | 分子名称: | 4-oxalocrotonate tautomerase, putative | | 著者 | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D. | | 登録日 | 2010-03-06 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
4S02
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, F42W, Y79A, and F123Y mutant | | 分子名称: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-30 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4RKJ
 
 | | Crystal structure of thrombin mutant S195T (free form) | | 分子名称: | GLYCEROL, POTASSIUM ION, Thrombin heavy chain, ... | | 著者 | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | 登録日 | 2014-10-13 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
3M21
 
 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | 分子名称: | Probable tautomerase HP_0924 | | 著者 | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | 登録日 | 2010-03-06 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
2WEG
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | 分子名称: | 2-fluorobenzenesulfonamide, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | 著者 | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | 登録日 | 2009-03-31 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2VWE
 
 | | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralizing Antibody Fab Fragment | | 分子名称: | ANTI-VEGF-B MONOCLONAL ANTIBODY, VASCULAR ENDOTHELIAL GROWTH FACTOR B | | 著者 | Leonard, P, Scotney, P.D, Jabeen, T, Iyer, S, Fabri, L.J, Nash, A.D, Acharya, K.R. | | 登録日 | 2008-06-23 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralising Antibody Fab Fragment.
J.Mol.Biol., 384, 2008
|
|
3MYZ
 
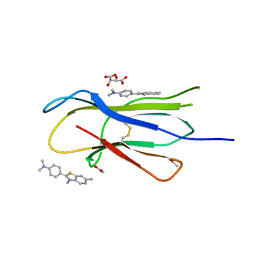 | | Protein induced photophysical changes to the amyloid indicator dye, thioflavin T | | 分子名称: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, ACETATE ION, Beta-2-microglobulin, ... | | 著者 | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | 登録日 | 2010-05-11 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MZT
 
 | | Protein-induced photophysical changes to the amyloid indicator dye, thioflavin T | | 分子名称: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Beta-2-microglobulin, COPPER (II) ION | | 著者 | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | 登録日 | 2010-05-13 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4RRN
 
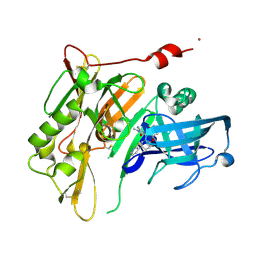 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4S,4a'S,10a'R)-2-amino-8'-(2-fluoropyridin-3-yl)-1-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2014-12-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RKO
 
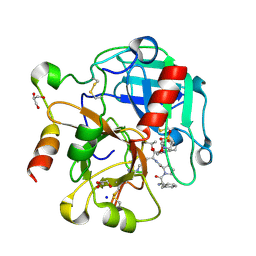 | | Crystal structure of thrombin mutant S195T bound with PPACK | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | 著者 | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | 登録日 | 2014-10-13 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
2VS7
 
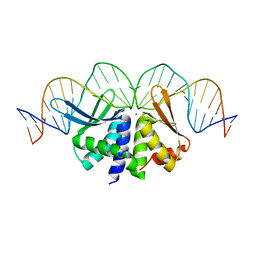 | | The crystal structure of I-DmoI in complex with DNA and Ca | | 分子名称: | 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*CP*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*G)-3', ACETATE ION, ... | | 著者 | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | 登録日 | 2008-04-21 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WEJ
 
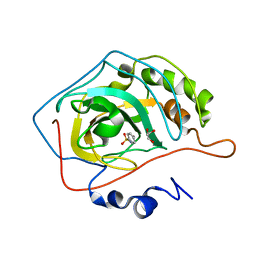 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | 分子名称: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION, ... | | 著者 | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | 登録日 | 2009-03-31 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
4S0I
 
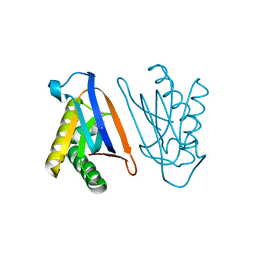 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123A mutant | | 分子名称: | Threonine--tRNA ligase | | 著者 | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | 登録日 | 2014-12-31 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
3NBK
 
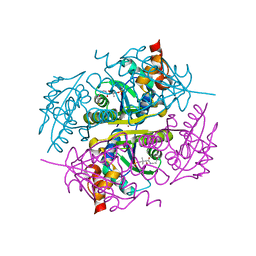 | |
4U1G
 
 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to monoclonal antibody QA1 | | 分子名称: | QA1 monoclonal antibody heavy chain, QA1 monoclonal antibody light chain, Reticulocyte binding protein 5 | | 著者 | Wright, K.E, Hjerrild, K.A, Bartlett, J, Douglas, A.D, Jin, J, Brown, R.E, Ashfield, R, Clemmensen, S.B, de Jongh, W.A, Draper, S.J, Higgins, M.K. | | 登録日 | 2014-07-15 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies.
Nature, 515, 2014
|
|
4U4V
 
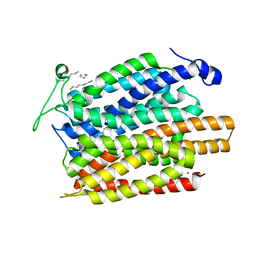 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | 分子名称: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | 著者 | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | 著者 | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
2VH0
 
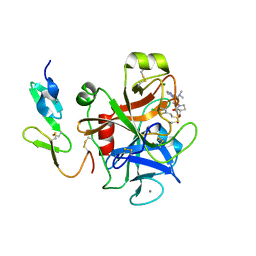 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | 分子名称: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | 著者 | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | 登録日 | 2007-11-16 | | 公開日 | 2008-11-25 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
3NBA
 
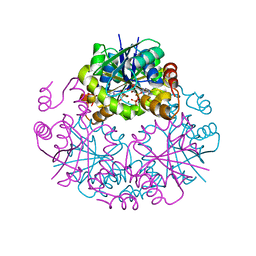 | | Phosphopantetheine Adenylyltranferase from Mycobacterium tuberculosis in complex with adenosine-5'-[(alpha,beta)-methyleno]triphosphate (AMPCPP) | | 分子名称: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Phosphopantetheine adenylyltransferase | | 著者 | Wubben, T.J, Mesecar, A.D. | | 登録日 | 2010-06-03 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Kinetic, thermodynamic, and structural insight into the mechanism of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis.
J.Mol.Biol., 404, 2010
|
|
