6WH4
 
 | | Crystal structure of HTR2A with inverse agonist | | 分子名称: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | 著者 | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | 登録日 | 2020-04-07 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6WHA
 
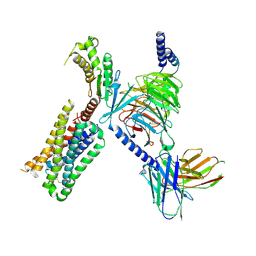 | | HTR2A bound to 25-CN-NBOH in complex with a mini-Galpha-q protein, beta/gamma subunits and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | 分子名称: | 4-(2-{[(2-hydroxyphenyl)methyl]amino}ethyl)-2,5-dimethoxybenzonitrile, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Skiniotis, G, Roth, B, Kim, K, Panova, O. | | 登録日 | 2020-04-07 | | 公開日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6WGT
 
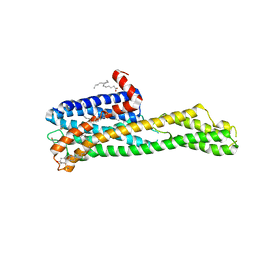 | | Crystal structure of HTR2A with hallucinogenic agonist | | 分子名称: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | 著者 | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | 登録日 | 2020-04-06 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
4XV5
 
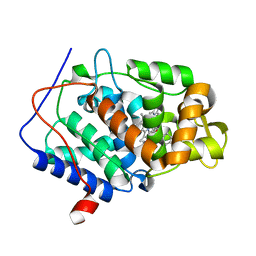 | | CcP gateless cavity | | 分子名称: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV7
 
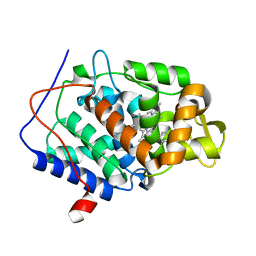 | | CcP gateless cavity | | 分子名称: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV8
 
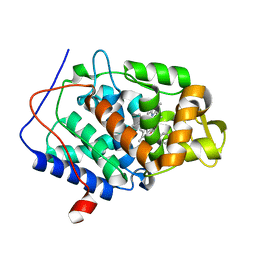 | | CcP gateless cavity | | 分子名称: | BENZAMIDINE, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV6
 
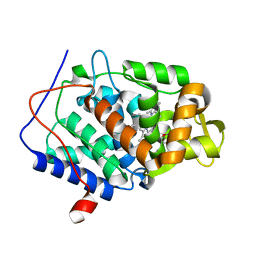 | | CcP gateless cavity | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XVA
 
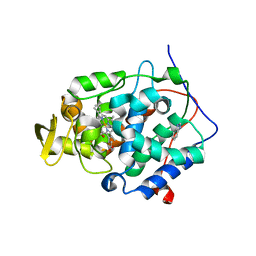 | |
4XV4
 
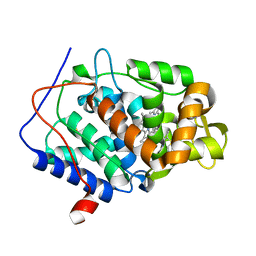 | | CcP gateless cavity | | 分子名称: | 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
189L
 
 | |
3BM6
 
 | | AmpC beta-lactamase in complex with a p.carboxyphenylboronic acid | | 分子名称: | 4-(dihydroxyboranyl)-2-({[4-(phenylsulfonyl)thiophen-2-yl]sulfonyl}amino)benzoic acid, Beta-lactamase | | 著者 | Tondi, D. | | 登録日 | 2007-12-12 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1TSL
 
 | |
1TSM
 
 | |
8UWL
 
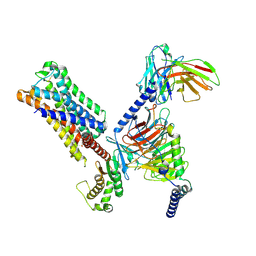 | | 5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | 分子名称: | 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | 登録日 | 2023-11-06 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
8V6U
 
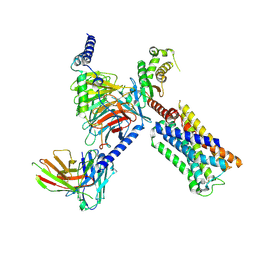 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | 分子名称: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | 著者 | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | 登録日 | 2023-12-03 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
6DPZ
 
 | |
6DPT
 
 | |
6DPY
 
 | |
8GLS
 
 | |
6DPX
 
 | |
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.77 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
