8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.35534 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
6EEB
 
 | | Calmodulin in complex with malbrancheamide | | 分子名称: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | 著者 | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | 登録日 | 2018-08-13 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3X21
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | 分子名称: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | 著者 | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | 登録日 | 2014-12-06 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.002 Å) | | 主引用文献 | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
8EE4
 
 | | Structure of PtuA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | 著者 | Shen, Z.F, Fu, T.M. | | 登録日 | 2022-09-06 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE7
 
 | |
8EEA
 
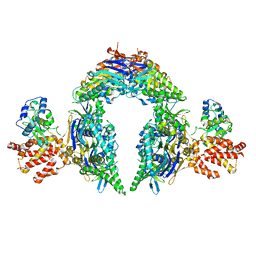 | | Structure of E.coli Septu (PtuAB) complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | 著者 | Shen, Z.F, Fu, T.M. | | 登録日 | 2022-09-06 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6M3M
 
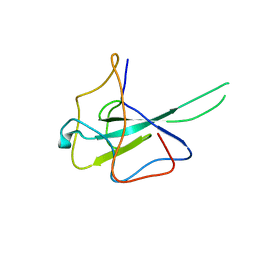 | |
6AAY
 
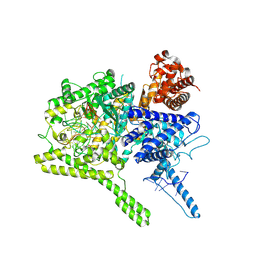 | | the Cas13b binary complex | | 分子名称: | Bergeyella zoohelcum Cas13b (R1177A) mutant, RNA (52-MER) | | 著者 | Bo, Z, Weiwei, Y, Yangmiao, Y, Ouyang, S.Y. | | 登録日 | 2018-07-19 | | 公開日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into Cas13b-guided CRISPR RNA maturation and recognition.
Cell Res., 28, 2018
|
|
7YTX
 
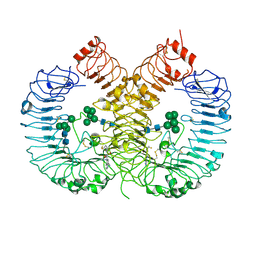 | | Crystal structure of TLR8 in complex with its antagonist | | 分子名称: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shimizu, T, Sakaniwa, K. | | 登録日 | 2022-08-16 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A novel Toll-like receptor 7/8-specific antagonist E6742 ameliorates clinically relevant disease parameters in murine models of lupus.
Eur.J.Pharmacol., 957, 2023
|
|
8AHH
 
 | |
8AHG
 
 | |
8AHI
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | 分子名称: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | 著者 | Baker, L.M, Murray, J.B, Hubbard, R.E. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | 分子名称: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | 著者 | Baker, L.M, Murray, J.B, Hubbard, R.E. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.271 Å) | | 主引用文献 | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8BT6
 
 | |
1U4A
 
 | | Solution structure of human SUMO-3 C47S | | 分子名称: | Ubiquitin-like protein SMT3A | | 著者 | Ding, H, Xu, Y, Dai, H, Tang, Y, Wu, J, Shi, Y. | | 登録日 | 2004-07-23 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Human SUMO-3 C47S and Its Binding Surface for Ubc9
Biochemistry, 44, 2005
|
|
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | 分子名称: | Esterase, PHOSPHATE ION | | 著者 | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | 登録日 | 2014-10-23 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
8T9Z
 
 | | Structural of M8C10 Fab in complex human metapneumovirus fusion protein | | 分子名称: | Fusion glycoprotein F0, M8C10 Fab Heavy Chain, M8C10 Fab Light Chain | | 著者 | Su, H.P, Eddins, M.J, Shipman, J.M, Kostas, J, Reid, J.C. | | 登録日 | 2023-06-26 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.995 Å) | | 主引用文献 | Structural characterization of M8C10, a neutralizing antibody targeting a highly conserved prefusion-specific epitope on the metapneumovirus fusion trimerization interface.
J.Virol., 97, 2023
|
|
8SUX
 
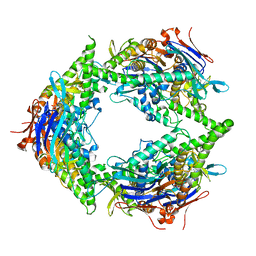 | |
4L07
 
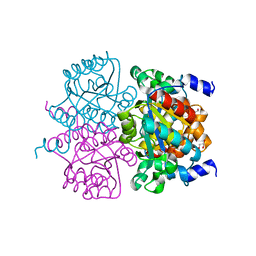 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | 分子名称: | GLYCEROL, Hydrolase, isochorismatase family, ... | | 著者 | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | 登録日 | 2013-05-30 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
6O9H
 
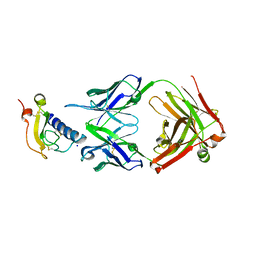 | | Mouse ECD with Fab1 | | 分子名称: | Gastric inhibitory polypeptide receptor, Heavy chain, Light chain, ... | | 著者 | Min, X, Wang, Z. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
