6BNE
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | 分子名称: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
6VU7
 
 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | 分子名称: | CHLORIDE ION, YbjN protein | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
6C5C
 
 | | Crystal structure of the 3-dehydroquinate synthase (DHQS) domain of Aro1 from Candida albicans SC5314 in complex with NADH | | 分子名称: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, CHLORIDE ION, ... | | 著者 | Michalska, K, Evdokimova, E, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-01-16 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
2IAI
 
 | | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3 | | 分子名称: | Putative transcriptional regulator SCO3833 | | 著者 | Zimmerman, M.D, Xu, X, Wang, S, Gu, J, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-09-08 | | 公開日 | 2006-09-26 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3
To be Published
|
|
6BNF
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | 分子名称: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
6WON
 
 | | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium | | 分子名称: | CHLORIDE ION, SULFATE ION, YohF | | 著者 | Stogios, P.J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-25 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium
To Be Published
|
|
6WOP
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | 分子名称: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-25 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
2HR3
 
 | | Crystal structure of putative transcriptional regulator protein from Pseudomonas aeruginosa PA01 at 2.4 A resolution | | 分子名称: | Probable transcriptional regulator | | 著者 | Kirillova, O, Chruszcz, M, Evdokimova, E, Kudritska, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-07-19 | | 公開日 | 2006-09-19 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of putative transcriptional regulator protein from Pseudomonas aeruginosa PA01 at 2.4 A resolution
To be Published
|
|
2FEX
 
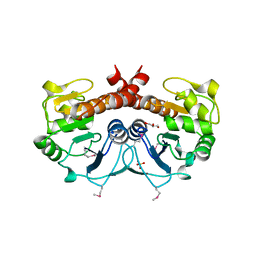 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | 分子名称: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | 著者 | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-16 | | 公開日 | 2006-01-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2FA1
 
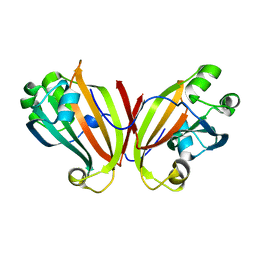 | | Crystal structure of PhnF C-terminal domain | | 分子名称: | Probable transcriptional regulator phnF, beta-D-fructopyranose | | 著者 | Lunin, V.V, Nocek, B.P, Gorelik, M, Skarina, T, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-06 | | 公開日 | 2006-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural characterization of GntR/HutC family signaling domain.
Protein Sci., 15, 2006
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | 分子名称: | Hypothetical protein AF2331 | | 著者 | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-14 | | 公開日 | 2006-01-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2FEF
 
 | | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa | | 分子名称: | 1,2-ETHANEDIOL, hypothetical protein PA2201 | | 著者 | Cymborowski, M.T, Chruszcz, M, Wang, S, Shumilin, I, Kudritska, M, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-15 | | 公開日 | 2006-01-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa
To be Published
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | 分子名称: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | 著者 | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | 登録日 | 2002-11-28 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
1OSC
 
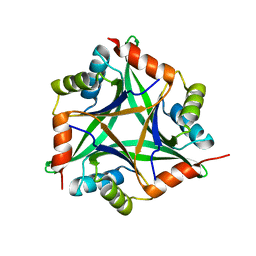 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | 分子名称: | similar to divalent cation tolerant protein CUTA | | 著者 | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-03-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
7KES
 
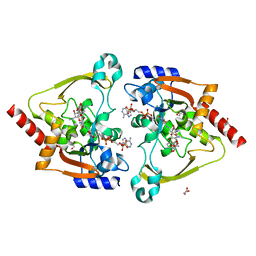 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | 分子名称: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-10-12 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
3ZQD
 
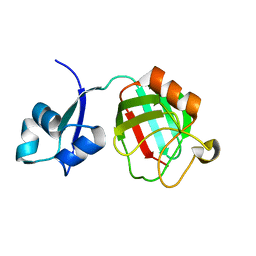 | | B. subtilis L,D-transpeptidase | | 分子名称: | L, D-TRANSPEPTIDASE YKUD | | 著者 | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | 登録日 | 2011-06-09 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
2I8E
 
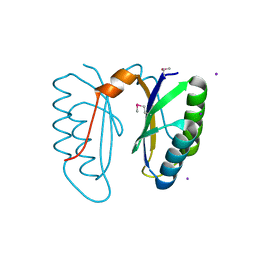 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | 分子名称: | Hypothetical protein, IODIDE ION | | 著者 | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-09-01 | | 公開日 | 2006-09-26 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
7LAP
 
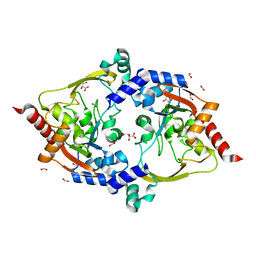 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Xa | | 分子名称: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | 著者 | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-01-06 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7LAO
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IIb | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside N(3)-acetyltransferase III, MAGNESIUM ION | | 著者 | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-01-06 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
1L5X
 
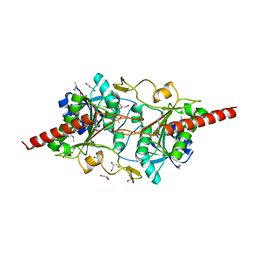 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | 分子名称: | ACETIC ACID, GLYCEROL, Survival protein E | | 著者 | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | 登録日 | 2002-03-08 | | 公開日 | 2003-02-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
4EXO
 
 | | Revised, rerefined crystal structure of PDB entry 2QHK, methyl accepting chemotaxis protein | | 分子名称: | Methyl-accepting chemotaxis protein, PYRUVIC ACID | | 著者 | Sweeney, E.G, Henderson, J.N, Goers, J, Wreden, C, Hicks, K.G, Foster, J.K, Parthasarathy, R, Remington, S.J, Guillemin, K. | | 登録日 | 2012-04-30 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Proposed Mechanism for the pH-Sensing Helicobacter pylori Chemoreceptor TlpB.
Structure, 20, 2012
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | 分子名称: | Effector protein hopAB3 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | 分子名称: | Effector protein hopAB1 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
6E1X
 
 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-07-10 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
3I1J
 
 | | Structure of a putative short chain dehydrogenase from Pseudomonas syringae | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-06-26 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a putative short chain dehydrogenase from Pseudomonas syringae
To be Published
|
|
