7DUP
 
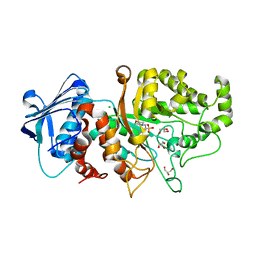 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
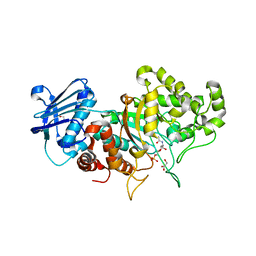 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
6CGI
 
 | | Structure of Salmonella Effector SseK3 | | 分子名称: | Type III secretion system effector protein, URIDINE-5'-DIPHOSPHATE | | 著者 | Chung, I.Y.W, Cygler, M. | | 登録日 | 2018-02-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Salmonella Effectors SseK1 and SseK3 Target Death Domain Proteins in the TNF and TRAIL Signaling Pathways.
Mol.Cell Proteomics, 18, 2019
|
|
2MKX
 
 | |
1D1Z
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP | | 分子名称: | SAP SH2 DOMAIN, SULFATE ION | | 著者 | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | 登録日 | 1999-09-22 | | 公開日 | 1999-10-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
1D4T
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP IN COMPLEX WITH A SLAM PEPTIDE | | 分子名称: | SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE, T CELL SIGNAL TRANSDUCTION MOLECULE SAP | | 著者 | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | 登録日 | 1999-10-06 | | 公開日 | 1999-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
1D4W
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP IN COMPLEX WITH SLAM PHOSPHOPEPTIDE | | 分子名称: | SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE, T CELL SIGNAL TRANSDUCTION MOLECULE SAP | | 著者 | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | 登録日 | 1999-10-06 | | 公開日 | 1999-10-14 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
6J8F
 
 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | 分子名称: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | 著者 | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2019-01-18 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.283 Å) | | 主引用文献 | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J8N
 
 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | 分子名称: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | 著者 | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2019-01-20 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J9H
 
 | |
6JOM
 
 | |
6J91
 
 | | Structure of a hypothetical protease | | 分子名称: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | 著者 | Liao, S, Gao, J, Xu, C. | | 登録日 | 2019-01-21 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
8TMA
 
 | |
8TM1
 
 | |
8WEX
 
 | |
7ENO
 
 | | Mutant strain M3 of foot-and-mouth disease virus type O | | 分子名称: | VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, VP3 of O type FMDV capsid, ... | | 著者 | Dong, H, Lu, Y. | | 登録日 | 2021-04-18 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
7ENP
 
 | | wild type of O type Foot-and-mouth disease virus | | 分子名称: | VP1 of O type FMDV capsid protein, VP2 of O type FMDV capsid protein, VP3 of O type FMDV capsid protein, ... | | 著者 | Dong, H, Lu, Y. | | 登録日 | 2021-04-18 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
7NS6
 
 | |
4MKI
 
 | | Cobalt transporter ATP-binding subunit | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | 著者 | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | 登録日 | 2013-09-05 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | 著者 | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2006-11-06 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
3R8B
 
 | |
8H4I
 
 | | DHA-bound FFAR4 in complex with Gs | | 分子名称: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | He, Y, Yin, H. | | 登録日 | 2022-10-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4L
 
 | | DHA-bound FFAR4 in complex with Gq | | 分子名称: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | He, Y, Yin, H. | | 登録日 | 2022-10-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4K
 
 | | GW9508-bound FFAR4 in complex with Gq | | 分子名称: | 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | He, Y, Yin, H. | | 登録日 | 2022-10-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
