7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | 分子名称: | D-Cysteine desulfhydrase | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | 分子名称: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7LCJ
 
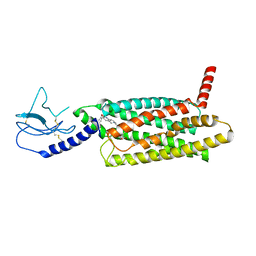 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | 分子名称: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | 著者 | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | 登録日 | 2021-01-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
7LCK
 
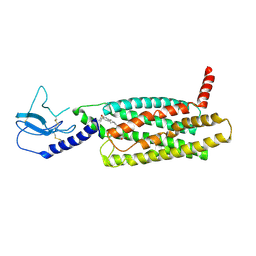 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R) | | 分子名称: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | 著者 | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | 登録日 | 2021-01-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
8HK1
 
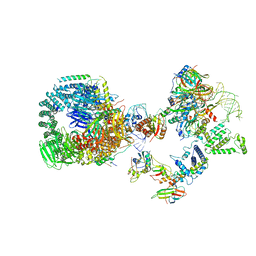 | | The cryo-EM structure of human pre-17S U2 snRNP | | 分子名称: | ATP-dependent RNA helicase DDX42, HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, ... | | 著者 | Zhang, X, Zhan, X, Shi, Y. | | 登録日 | 2022-11-24 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly.
Nat Commun, 14, 2023
|
|
8INB
 
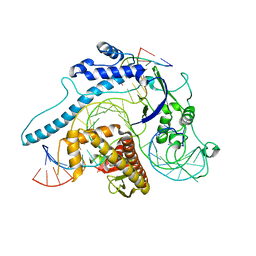 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | 分子名称: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | 著者 | Zhang, X, Duan, Z.Q, Zhu, J.K. | | 登録日 | 2023-03-09 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
8WRQ
 
 | |
7VH2
 
 | |
7VGQ
 
 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Zhang, X, Ma, J, Zhang, S. | | 登録日 | 2021-09-18 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH1
 
 | | Cryo-EM structure of Machupo virus dimeric L-Z complex | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Zhang, X, Ma, J, Zhang, S. | | 登録日 | 2021-09-20 | | 公開日 | 2021-09-29 | | 最終更新日 | 2025-03-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7WOP
 
 | | The local refined map of Omicron spike with bispecific antibody FD01 | | 分子名称: | 16L9, GW01, Spike protein S1 | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOR
 
 | | The state 2 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOV
 
 | | The state 5 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | 著者 | Zhang, X, Zhan, W.Q, Sun, L, Chen, Z.G. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOS
 
 | | The state 3 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOW
 
 | | The state 6 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, GW01 Fv, Spike glycoprotein | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (6.11 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOU
 
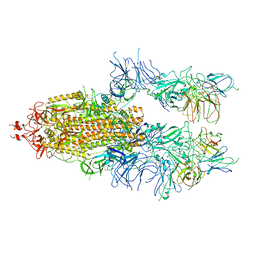 | | The state 4 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOQ
 
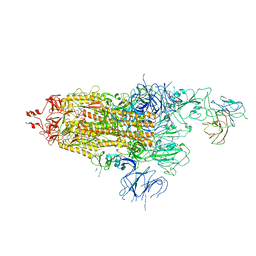 | | The state 1 of Omicron Spike with bispecific antibody FD01 | | 分子名称: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
8K52
 
 | | Cryo-EM structure of chitin synthase | | 分子名称: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | 著者 | Zhang, X, Niu, S, Li, P, Bi, Y. | | 登録日 | 2023-07-20 | | 公開日 | 2024-12-04 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | 分子名称: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | 著者 | Zhang, X, Ma, J, Chen, L. | | 登録日 | 2018-10-23 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
7DY9
 
 | | Thermotoga maritima ferritin mutant-FLAL | | 分子名称: | Ferritin | | 著者 | Zhao, G, Zhang, X. | | 登録日 | 2021-01-20 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DYB
 
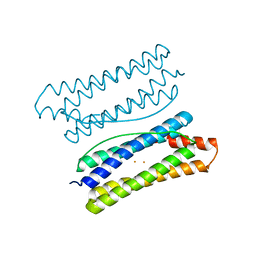 | | Thermotoga maritima ferritin mutant-FLAL-L | | 分子名称: | FE (III) ION, Ferritin | | 著者 | Zhao, G, Zhang, X. | | 登録日 | 2021-01-20 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.779 Å) | | 主引用文献 | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
8Z0O
 
 | | Cryo-EM structure of chitin synthase | | 分子名称: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | 著者 | Zhang, X, Niu, S, Li, P, Bi, Y. | | 登録日 | 2024-04-10 | | 公開日 | 2024-12-04 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
7CTH
 
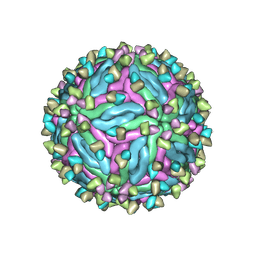 | | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, Single Chain Variable Fragment | | 著者 | Zhang, X, Sharma, A, Duquerroy, S, Zhou, Z.H, Rey, F.A. | | 登録日 | 2020-08-19 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7V66
 
 | | Structure of Apoferritin | | 分子名称: | Ferritin heavy chain | | 著者 | Zhang, X, Wu, C, Shi, H. | | 登録日 | 2021-08-19 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (1.89 Å) | | 主引用文献 | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
7X5B
 
 | | Crystal structure of RuvB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
