1IVF
 
 | |
7DW7
 
 | | Crystal Structure of N1051A mutant of Formylglycinamidine Synthetase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Sharma, N, Tanwar, A.S, Anand, R. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
7KXJ
 
 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | 著者 | Li, Z, Rini, J. | | 登録日 | 2020-12-04 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
5INJ
 
 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-03-07 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | 分子名称: | CalU16 | | 著者 | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-22 | | 公開日 | 2012-12-12 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
2IIT
 
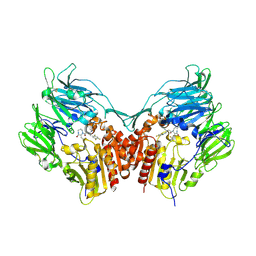 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | 分子名称: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Biftu, T, Weber, A.E. | | 登録日 | 2006-09-28 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2IIV
 
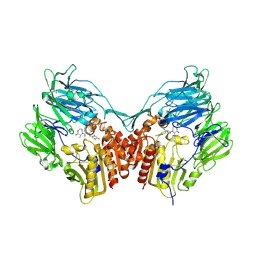 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | 分子名称: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-METHYL-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Weber, A.E, Biftu, T. | | 登録日 | 2006-09-28 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3D4L
 
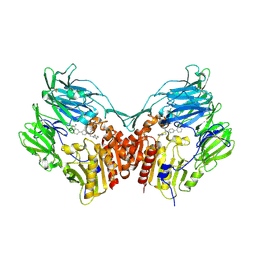 | | Human dipeptidyl peptidase IV/CD26 in complex with a novel inhibitor | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G. | | 登録日 | 2008-05-14 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of new binding elements in DPP-4 inhibition and their applications in novel DPP-4 inhibitor design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2P8S
 
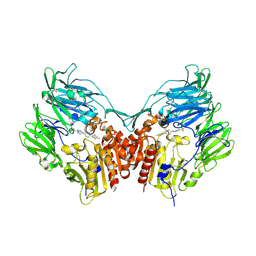 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | 分子名称: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Biftu, T. | | 登録日 | 2007-03-23 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | 分子名称: | BROMIDE ION, putative acetyltransferase | | 著者 | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2004-10-04 | | 公開日 | 2004-10-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
5Z5W
 
 | |
5Z5X
 
 | |
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | 分子名称: | COENZYME A, Protein At1g77540 | | 著者 | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-10-02 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.054 Å) | | 主引用文献 | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
6LFH
 
 | |
6LYM
 
 | | Crystal structure of D657A mutant of formylglycinamidine synthetase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Sharma, N, Tanwar, A.S, Anand, R. | | 登録日 | 2020-02-14 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYO
 
 | | Crystal Structure of H296A mutant of Formylglycinamidine Synthetase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Sharma, N, Tanwar, A.S, Anand, R. | | 登録日 | 2020-02-15 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYK
 
 | | Crystal Structure of R1263A mutant of Formylglycinamidine Synthetase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | 著者 | Sharma, N, Tanwar, A.S, Anand, R. | | 登録日 | 2020-02-14 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYL
 
 | | Crystal structure of S1052D mutant of Formylglycinamidine synthetase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Sharma, N, Tanwar, A.S, Anand, R. | | 登録日 | 2020-02-14 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
3D0R
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group P2(1) | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein CalG3 | | 著者 | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | 登録日 | 2008-05-02 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | 分子名称: | PHOSPHATE ION, Tryptophan halogenase | | 著者 | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | 登録日 | 2006-12-14 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2NAT
 
 | |
2NAU
 
 | |
1OVE
 
 | | The structure of p38 alpha in complex with a dihydroquinolinone | | 分子名称: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERIDIN-4-YL-3,4-DIHYDROQUINOLIN-2(1H)-ONE, GLYCEROL, Mitogen-activated protein kinase 14 | | 著者 | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | 登録日 | 2003-03-26 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
3CTM
 
 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | 分子名称: | Carbonyl Reductase | | 著者 | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | 登録日 | 2008-04-14 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
4NJE
 
 | | Crystal structure of Pyrococcus furiosus L-asparaginase with ligand | | 分子名称: | ASPARTIC ACID, L-asparaginase | | 著者 | Sharma, P, Tomar, R, Singh, S, Yadav, S.P.S, Ashish, Kundu, B. | | 登録日 | 2013-11-09 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
