4LGW
 
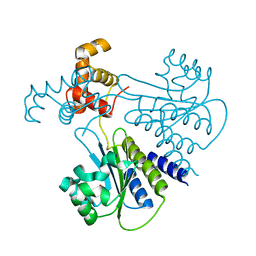 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | 分子名称: | GLYCEROL, Regulatory protein SdiA | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-28 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LFU
 
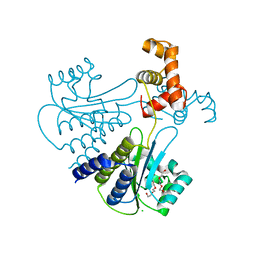 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | 分子名称: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-27 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P24
 
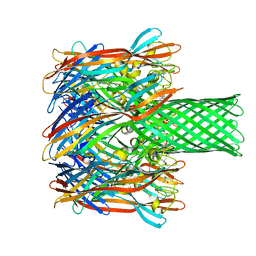 | | pore forming toxin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-hemolysin | | 著者 | Sugawara, T, Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | 登録日 | 2014-03-01 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for pore-forming mechanism of staphylococcal alpha-hemolysin.
Toxicon, 108, 2015
|
|
2P4L
 
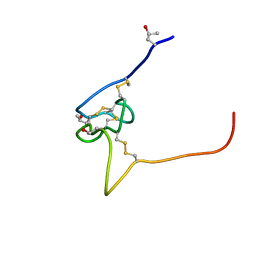 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | 分子名称: | I-superfamily conotoxin r11a | | 著者 | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | 登録日 | 2007-03-12 | | 公開日 | 2007-09-25 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
7XAR
 
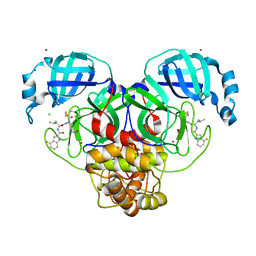 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | 分子名称: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | 著者 | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | 登録日 | 2022-03-18 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
3AK4
 
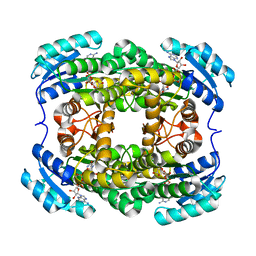 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | 分子名称: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | 登録日 | 2010-07-07 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
1WRI
 
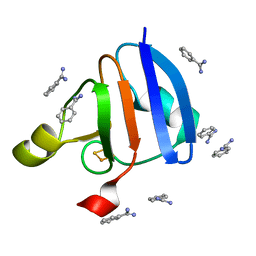 | | Crystal Structure of Ferredoxin isoform II from E. arvense | | 分子名称: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin II | | 著者 | Kurisu, G, Nishiyama, D, Kusunoki, M, Fujikawa, S, Katoh, M, Hanke, G.T, Hase, T, Teshima, K. | | 登録日 | 2004-10-18 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A structural basis of Equisetum arvense ferredoxin isoform II producing an alternative electron transfer with ferredoxin-NADP+ reductase.
J.Biol.Chem., 280, 2005
|
|
3AQK
 
 | |
3AQN
 
 | |
3AQL
 
 | |
3AQM
 
 | |
5KKY
 
 | |
6RYA
 
 | |
6RYB
 
 | |
4MZ9
 
 | | Revised structure of E. coli SSB | | 分子名称: | Single-stranded DNA-binding protein | | 著者 | Oakley, A.J. | | 登録日 | 2013-09-29 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy.
Nucleic Acids Res., 42, 2014
|
|
6AE1
 
 | |
6AE2
 
 | |
6LJA
 
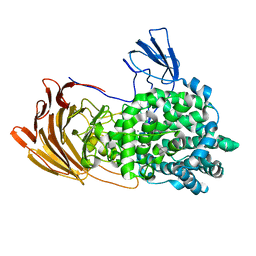 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | 著者 | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | 登録日 | 2019-12-13 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6LJL
 
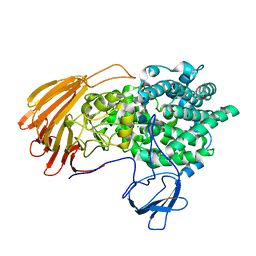 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | 分子名称: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | 著者 | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | 分子名称: | Alpha-synuclein | | 著者 | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | 登録日 | 2014-10-06 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.854 Å) | | 主引用文献 | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
5K2H
 
 | |
5K2F
 
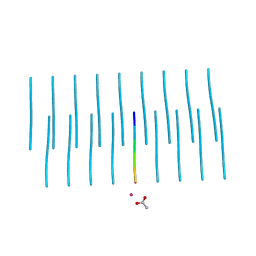 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | 分子名称: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | 著者 | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1 Å) | | 主引用文献 | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | 分子名称: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | 著者 | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1 Å) | | 主引用文献 | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
7OY6
 
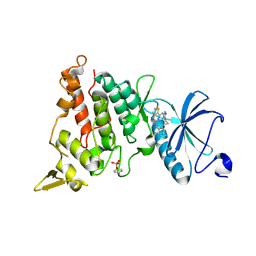 | | Crystal structure of human DYRK1A in complex with ARN25068 | | 分子名称: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | 著者 | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | 登録日 | 2021-06-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
