7SQ1
 
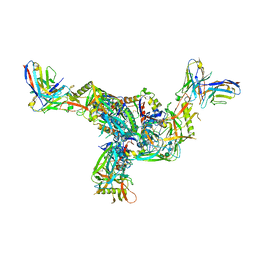 | | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C05 Fab Light chain, ... | | 著者 | Moore, A, Du, J, Xu, Z, Walker, S, Kulp, D.W, Pallesen, J. | | 登録日 | 2021-11-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer.
Nat Commun, 13, 2022
|
|
5KSC
 
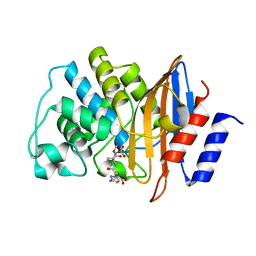 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | 著者 | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2019-12-04 | | 実験手法 | NEUTRON DIFFRACTION (2.1 Å) | | 主引用文献 | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
3KHI
 
 | |
8QUV
 
 | | Crystal structure of chlorite dismutase at 3000 eV with no absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QVB
 
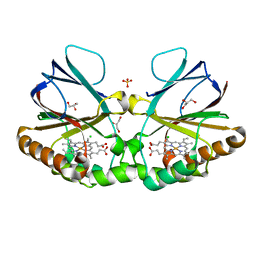 | | Crystal structure of chlorite dismutase at 3000 eV based on a combination of spherical harmonics and analytical absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUU
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on spherical harmonics absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUZ
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on analytical absorption corrections | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Duman, R, Wagner, A, Kamps, J, Orville, A. | | 登録日 | 2023-10-17 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
3UG9
 
 | | Crystal Structure of the Closed State of Channelrhodopsin | | 分子名称: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | 著者 | Kato, H.E, Ishitani, R, Nureki, O. | | 登録日 | 2011-11-02 | | 公開日 | 2012-01-25 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
3BQC
 
 | | High pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | 分子名称: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | 著者 | Niefind, K, Raaf, J, Issinger, O.-G. | | 登録日 | 2007-12-20 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
6ZWO
 
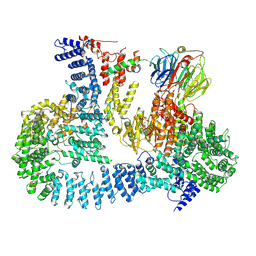 | | cryo-EM structure of human mTOR complex 2, focused on one half | | 分子名称: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | 登録日 | 2020-07-28 | | 公開日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6ZWM
 
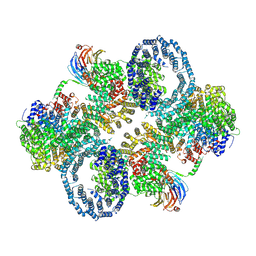 | | cryo-EM structure of human mTOR complex 2, overall refinement | | 分子名称: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | 登録日 | 2020-07-28 | | 公開日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
5UMK
 
 | |
5UMJ
 
 | |
4KF7
 
 | |
4KF8
 
 | |
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-18 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5YNY
 
 | | Structure of house dust mite allergen Der F 21 in PEG2KMME | | 分子名称: | Allergen Der f 21 | | 著者 | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I. | | 登録日 | 2017-10-25 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | 著者 | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-09 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | 分子名称: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | 著者 | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-15 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | 分子名称: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | 著者 | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | 分子名称: | D-dopachrome decarboxylase, SULFATE ION | | 著者 | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | 登録日 | 2021-05-09 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
4HKO
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | 登録日 | 2012-10-15 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
