5I4V
 
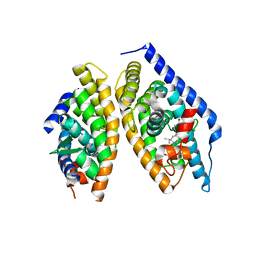 | | Discovery of novel, orally efficacious Liver X Receptor (LXR) beta agonists | | 分子名称: | Oxysterols receptor LXR-beta,Nuclear receptor coactivator 2, Retinoic acid receptor RXR-beta,Nuclear receptor coactivator 2, {2-[(2R)-4-[4-(hydroxymethyl)-3-(methylsulfonyl)phenyl]-2-(propan-2-yl)piperazin-1-yl]-4-(trifluoromethyl)pyrimidin-5-yl}methanol | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-02-12 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Discovery of a Novel, Orally Efficacious Liver X Receptor (LXR) beta Agonist.
J.Med.Chem., 59, 2016
|
|
2FJ2
 
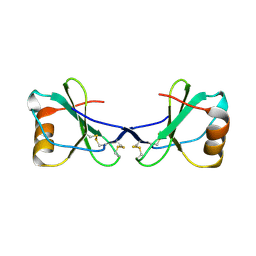 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | 分子名称: | Viral macrophage inflammatory protein-II | | 著者 | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | 登録日 | 2005-12-30 | | 公開日 | 2006-12-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2FHT
 
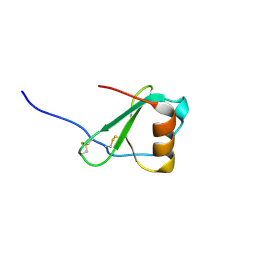 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | 分子名称: | Viral macrophage inflammatory protein-II | | 著者 | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | 登録日 | 2005-12-27 | | 公開日 | 2006-12-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2JKC
 
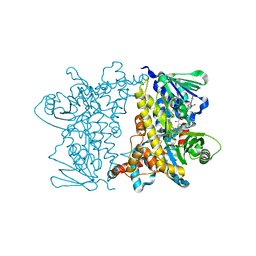 | |
8Y3X
 
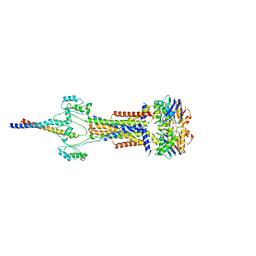 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | 著者 | Zhang, Z, Dong, H, Chen, Y. | | 登録日 | 2024-01-29 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | 著者 | Liu, Z, Yan, A, Gao, Y. | | 登録日 | 2022-10-20 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
6NFX
 
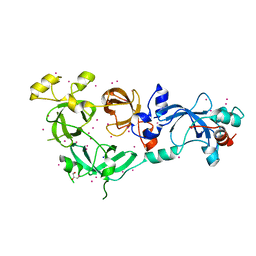 | | MBTD1 MBT repeats | | 分子名称: | GLYCEROL, MBT domain-containing protein 1,Enhancer of polycomb homolog 1, SODIUM ION, ... | | 著者 | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2018-12-21 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex.
Cell Rep, 30, 2020
|
|
8X61
 
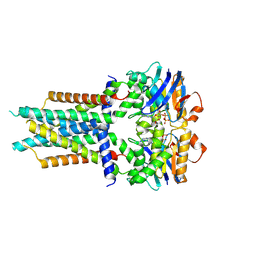 | | Cryo-EM structure of ATP-bound FtsE(E163Q)X | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX | | 著者 | Zhang, Z.Y, Chen, Y.T. | | 登録日 | 2023-11-20 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
8HFQ
 
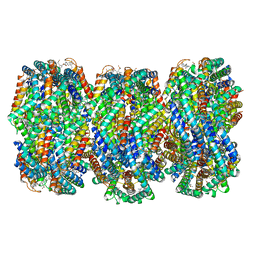 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | 分子名称: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | 著者 | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | 登録日 | 2022-11-11 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
5KYJ
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7F9W
 
 | | CD25 in complex with Fab | | 分子名称: | Heavy chain of Fab, Interleukin-2 receptor subunit alpha, Light chain of Fab | | 著者 | Liu, C. | | 登録日 | 2021-07-05 | | 公開日 | 2022-01-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity.
Sci Rep, 11, 2021
|
|
6PER
 
 | | Crystal Structure of Ligand-Free iSeroSnFR | | 分子名称: | 1,2-ETHANEDIOL, iSeroSnFR, a soluble, ... | | 著者 | Hartanto, S, Tian, L, Fisher, A.J. | | 登録日 | 2019-06-20 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning.
Cell, 183, 2020
|
|
4L6Q
 
 | | ROCK2 in complex with benzoxaborole | | 分子名称: | 6-[4-(aminomethyl)-2-fluorophenoxy]-2,1-benzoxaborol-1(3H)-ol, Rho-associated protein kinase 2 | | 著者 | Rock, F, Jarnagin, K. | | 登録日 | 2013-06-12 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Linking phenotype to kinase: identification of a novel benzoxaborole hinge-binding motif for kinase inhibition and development of high-potency rho kinase inhibitors.
J.Pharmacol.Exp.Ther., 347, 2013
|
|
7KJZ
 
 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | 分子名称: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | 著者 | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | 登録日 | 2020-10-26 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KJO
 
 | |
7KK7
 
 | | crystal structure of ligand-free PLEKHA7 PH domain | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | 著者 | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | 登録日 | 2020-10-27 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
3O0J
 
 | | PDE4B In complex with ligand an2898 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)oxy]benzene-1,2-dicarbonitrile, MAGNESIUM ION, ... | | 著者 | Alley, M.R.K, Zhou, Y. | | 登録日 | 2010-07-19 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Boron-based phosphodiesterase inhibitors show novel binding of boron to PDE4 bimetal center.
Febs Lett., 586, 2012
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | 分子名称: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
6L9J
 
 | | Structure of yeast Snf5 and Swi3 subcomplex | | 分子名称: | GLYCEROL, SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF complex subunit SWI3 | | 著者 | Long, J, Zhou, H. | | 登録日 | 2019-11-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.642 Å) | | 主引用文献 | Snf5 and Swi3 subcomplex formation is required for SWI/SNF complex function in yeast.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
7EP2
 
 | |
7EP1
 
 | |
7EP4
 
 | |
7EP5
 
 | |
