2PCX
 
 | |
5XFF
 
 | |
5Z78
 
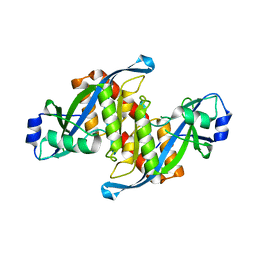 | | Structure of TIRR/53BP1 complex | | 分子名称: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | 著者 | Dai, Y.X, Shan, S. | | 登録日 | 2018-01-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR
Nat Commun, 9, 2018
|
|
8TVS
 
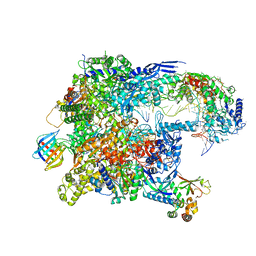 | | Cryo-EM structure of backtracked Pol II in complex with Rad26 | | 分子名称: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVQ
 
 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | 分子名称: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | 分子名称: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TUG
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | 分子名称: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-16 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | 分子名称: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | 分子名称: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | 分子名称: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | 分子名称: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8IMY
 
 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, T, Xu, Y, Qu, Q, Li, D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, Y, Li, T, Qu, Q, Li, D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8K9R
 
 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | 分子名称: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | 著者 | Li, T, Hong, J, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9Q
 
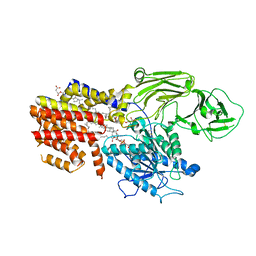 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | 著者 | Hong, J, Li, T, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9T
 
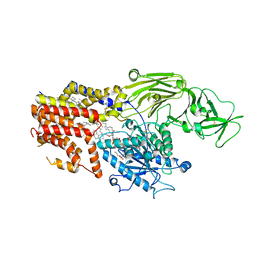 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | 分子名称: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | 著者 | Li, T, Hong, J, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
6N7I
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (gp4(5)-DNA) | | 分子名称: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | 登録日 | 2018-11-27 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7T
 
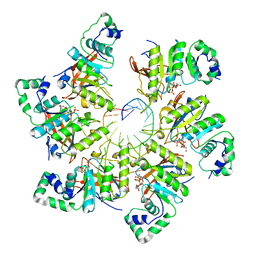 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form III) | | 分子名称: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | 登録日 | 2018-11-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7N
 
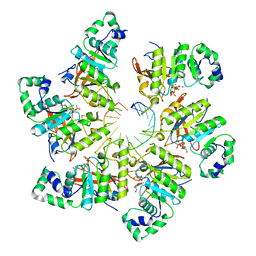 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form I) | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase/helicase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | 登録日 | 2018-11-27 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N9W
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS2) | | 分子名称: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | 登録日 | 2018-12-04 | | 公開日 | 2019-03-06 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7S
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form II) | | 分子名称: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | 著者 | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | 登録日 | 2018-11-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
4JQH
 
 | |
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
