2O3J
 
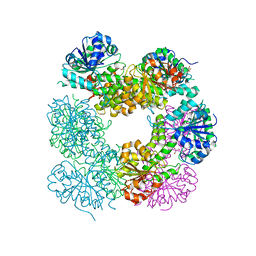 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
8S55
 
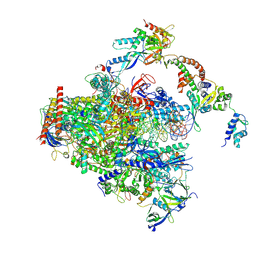 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
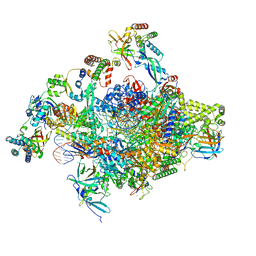 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S52
 
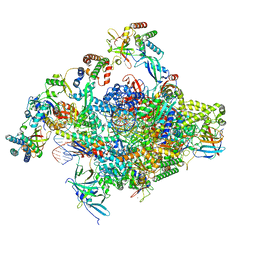 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S5N
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S54
 
 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling
J.Virol., 2024
|
|
2ABS
 
 | | Crystal structure of T. gondii adenosine kinase complexed with AMP-PCP | | Descriptor: | CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-16 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of Toxoplasma gondii adenosine kinase in complex with an ATP analog at 1.1 angstroms resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2A8Y
 
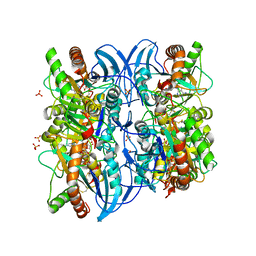 | | Crystal structure of 5'-deoxy-5'methylthioadenosine phosphorylase complexed with 5'-deoxy-5'methylthioadenosine and sulfate | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5'-methylthioadenosine phosphorylase (mtaP), SULFATE ION | | Authors: | Zhang, Y, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2005-07-10 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of 5'-deoxy-5'-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds.
J.Mol.Biol., 357, 2006
|
|
2A9Z
 
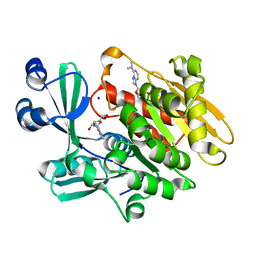 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine and AMP-PCP | | Descriptor: | 6N-DIMETHYLADENOSINE, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-12 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2A9Y
 
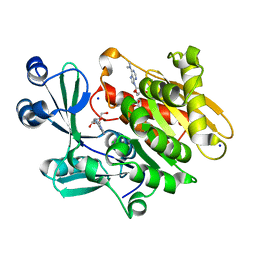 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine | | Descriptor: | 6N-DIMETHYLADENOSINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-12 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5BW0
 
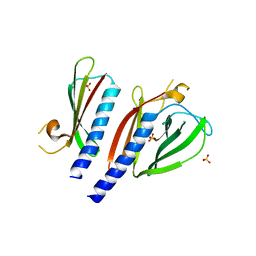 | | The crystal structure of minor pseudopilin binary complex of XcpV and XcpW from the Type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | SULFATE ION, Type II secretion system protein I, Type II secretion system protein J | | Authors: | Zhang, Y, Faucher, F, Poole, K, Jia, Z. | | Deposit date: | 2015-06-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
PLoS Pathog., 14, 2018
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
2KNQ
 
 | | Solution structure of E.Coli GspH | | Descriptor: | General secretion pathway protein H | | Authors: | Zhang, Y, Jin, C. | | Deposit date: | 2009-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GspH
To be Published
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
5M5G
 
 | | Crystal structure of the Chaetomium Thermophilum polycomb repressive complex 2 (PRC2) | | Descriptor: | Fragment from molecular 2 (region containing putative polycomb protein Suz12), HISTONE H3 11-Mer peptide, Putative uncharacterized protein, ... | | Authors: | Zhang, Y, Justin, N, Wilson, J, Gamblin, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
2AA0
 
 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AB8
 
 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside and AMP-PCP | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2A7Q
 
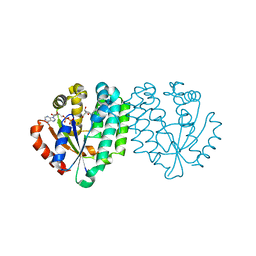 | | Crystal structure of human dCK complexed with clofarabine and ADP | | Descriptor: | 2-CHLORO-9-(2-DEOXY-2-FLUORO-B -D-ARABINOFURANOSYL)-9H-PURIN-6-AMINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, ... | | Authors: | Zhang, Y, Secrist III, J.A, Ealick, S.E. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
9CII
 
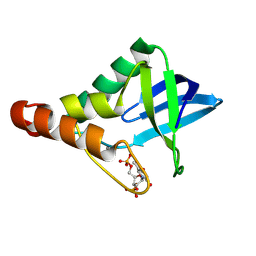 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Robinson, A.C, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature
To Be Published
|
|
9CIL
 
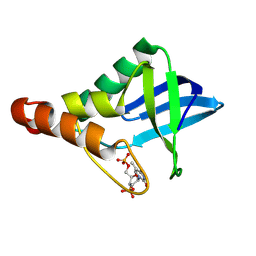 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature
To Be Published
|
|
9CIK
 
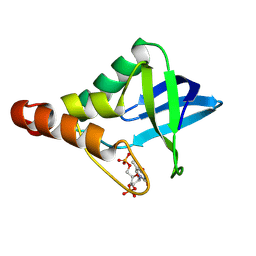 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, SIegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature
To Be Published
|
|
9CIJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, L.J, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature
To Be Published
|
|
6O7G
 
 | |
