2MPK
 
 | |
6P49
 
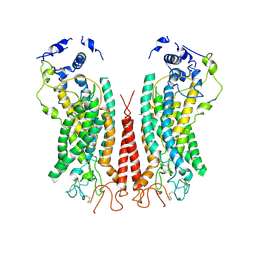 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P46
 
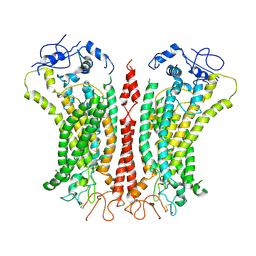 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
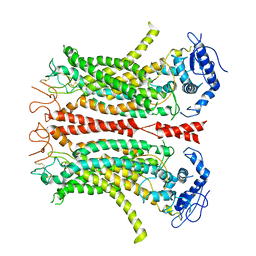 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P48
 
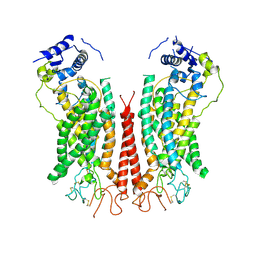 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
7B7O
 
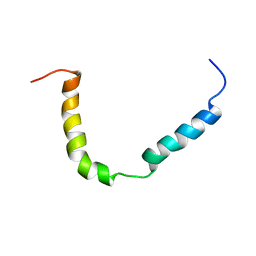 | | Solution structure of A. thaliana core TatA in DHPC micelles | | Descriptor: | Sec-independent protein translocase protein TATA, chloroplastic | | Authors: | Pettersson, P, Ye, W, Jakob, M, Tannert, F, Klosgen, R.B, Maler, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR.
FEBS J, 285, 2018
|
|
6BGJ
 
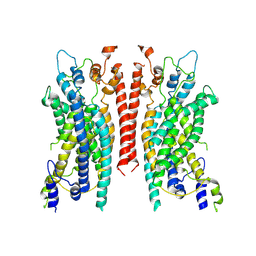 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
4G1T
 
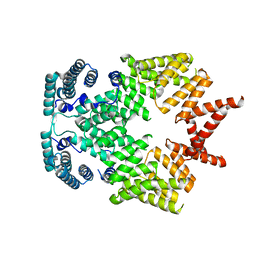 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
4MEJ
 
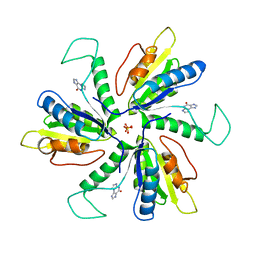 | | Crystal structure of Lactobacillus helveticus purine deoxyribosyl transferase (PDT) with the tricyclic purine 8,9-dihydro-9-oxoimidazo[2,1-b]purine (N2,3-ethenoguanine) | | Descriptor: | 3H-imidazo[2,1-b]purin-4(5H)-one, Nucleoside deoxyribosyltransferase, SULFATE ION | | Authors: | Paul, D, Seckute, J, Ealick, S.E. | | Deposit date: | 2013-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylation of a tricyclic purine analog at alternative
sites by nucleoside 2 -deoxyribosyltransferases
Plos One, 2014
|
|
8QNV
 
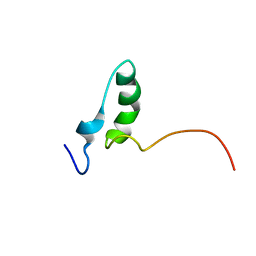 | |
8QNJ
 
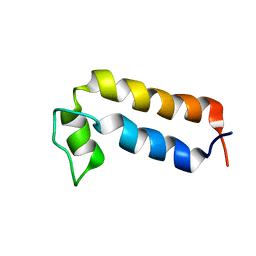 | |
8QNT
 
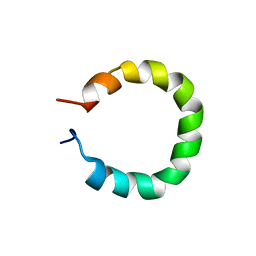 | |
7UKH
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, intracellular region | | Descriptor: | CALCIUM ION, Isoform 2 of Kv channel-interacting protein 2, Potassium voltage-gated channel subfamily D member 2, ... | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKE
 
 | |
7UK5
 
 | |
7UKG
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, transmembrane region | | Descriptor: | Dipeptidyl-peptidase 6, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKD
 
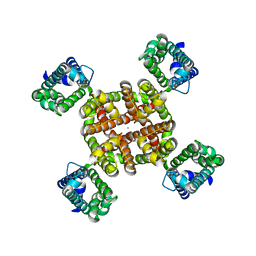 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
3PNW
 
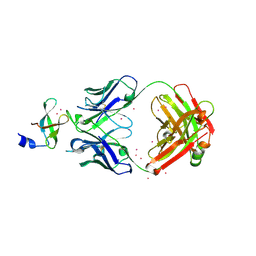 | | Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB | | Descriptor: | FAB heavy chain, FAB light chain, Tudor domain-containing protein 3, ... | | Authors: | Loppnau, P, Tempel, W, Wernimont, A.K, Lam, R, Ravichandran, M, Adams-Cioaba, M.A, Persson, H, Sidhu, S.S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
J.Mol.Biol., 425, 2013
|
|
6IV8
 
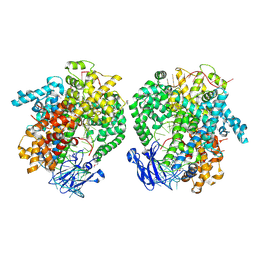 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
6IV9
 
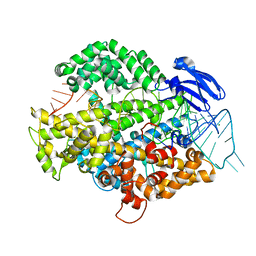 | | the Cas13d binary complex | | Descriptor: | Cas13d, MAGNESIUM ION, crRNA (50-MER) | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
6J8L
 
 | |
8CX9
 
 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
