4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7D58
 
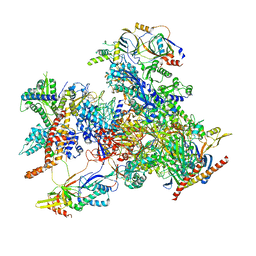 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
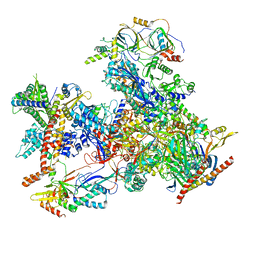 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5ZO0
 
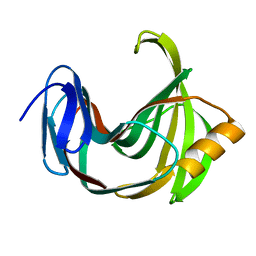 | | Neutron structure of xylanase at pD5.4 | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Wan, Q, Li, Z.H. | | Deposit date: | 2018-04-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.648 Å) | | Cite: | Neutron structure of xylanase at pD5.4
To be published
|
|
3NVO
 
 | |
3NWI
 
 | |
4PDJ
 
 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7D3Z
 
 | |
7D4L
 
 | |
7D4X
 
 | |
7D6G
 
 | |
7D49
 
 | |
9LXO
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9LXN
 
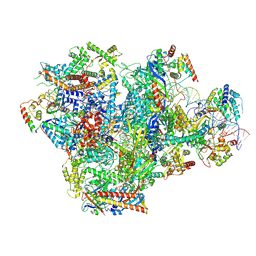 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
5XYB
 
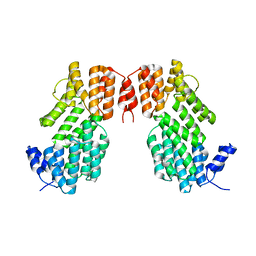 | |
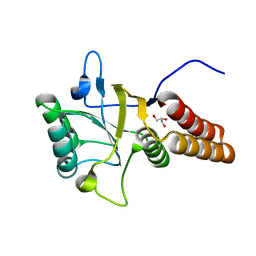
1FY9
 
 | |
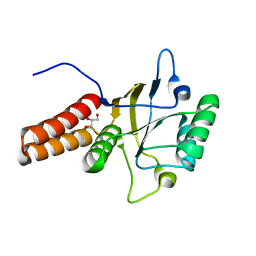
1FYA
 
 | |
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
9IMR
 
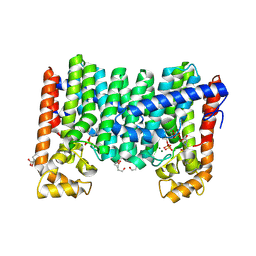 | | Crystal structure of geranylgeranyl pyrophosphate synthase Rv0562 from Mycobacterium tuberculosis in complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMQ
 
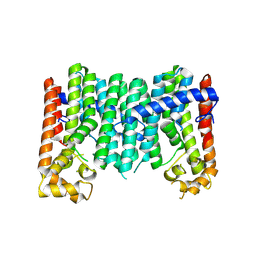 | | Crystal structure of a C45 isoprenyl diphosphate synthase, Rv0562 from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, Nonaprenyl diphosphate synthase | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMS
 
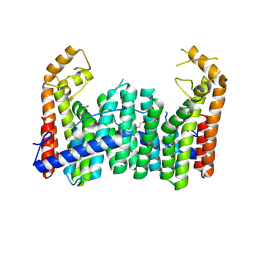 | |
