1H96
 
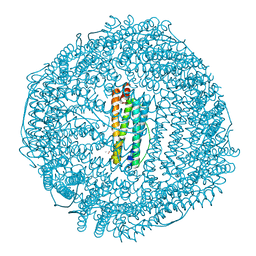 | | recombinant mouse L-chain ferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, SULFATE ION | | Authors: | Granier, T, Gallois, B, D'Estaintot, B.L, Dautant, A, Chevalier, J.M, Mellado, J.M, Beaumont, C, Santambrogio, P, Arosio, P, Precigoux, G. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mouse L-Chain Ferritin at 1.6 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3BS7
 
 | |
8XX0
 
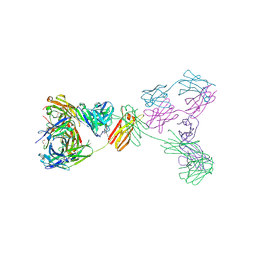 | | Crystal structure of anti-IgE antibody HMK-12 Fab complexed with IgE F(ab')2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SPE7 immunoglobulin E F(ab')2 heavy chain, ... | | Authors: | Hirano, T, Koyanagi, A, Kasai, M, Okumura, K. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric inhibition of IgE-Fc epsilon RI interactions by simultaneous targeting of IgE F(ab')2 epitopes.
Commun Biol, 7, 2024
|
|
4KLY
 
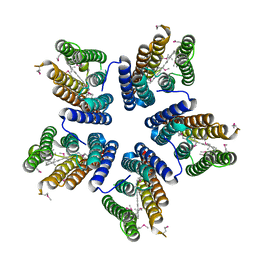 | | Crystal structure of a blue-light absorbing proteorhodopsin mutant D97N from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KNF
 
 | | Crystal structure of a blue-light absorbing proteorhodopsin double-mutant D97N/Q105L from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
7PUF
 
 | | urate oxidase azaxanthine complex under 600 bar (60 MPa) of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
5Y97
 
 | | Crystal structure of snake gourd seed lectin in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Seed lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Chandran, T, Vijayan, M, Sivaji, N, Surolia, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and retention in snake gourd seed lectin (SGSL). A crystallographic, thermodynamic and molecular dynamics study
Glycobiology, 28, 2018
|
|
6Q8S
 
 | |
6Q8R
 
 | |
6Q8Q
 
 | |
6QAR
 
 | |
5LX5
 
 | | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782-RP. | | Descriptor: | DIPHOSPHATE, Nicotinamide phosphoribosyltransferase, [(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-5-[[[4-[6-(ethylamino)-5-(2-piperidin-1-ylethylcarbamoyl)pyridin-2-yl]-2-fluoranyl-phenyl]carbamoylamino]methyl]pyridin-1-ium-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Bertrand, T, Marquette, J.P. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782-RP
To Be Published
|
|
5Y42
 
 | |
2PLO
 
 | |
6RA8
 
 | |
6RGM
 
 | | urate oxidase under 130 bar of krypton | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ACETATE ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
2LWB
 
 | | Structural model of BAD-1 repeat loop by NMR | | Descriptor: | Adhesin WI-1 | | Authors: | Brandhorst, T, Klein, B, Tonelli, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a fungal adhesin that mimics thrombospondin-1 by binding heparin sulfate glycosaminoglycan and suppressing T cell activation via interaction with CD47
To be Published
|
|
7PWN
 
 | | URATE OXYDASE AZA-XANTHINE COMPLEX AT 1000 BARS (100 MPa) OF ARGON | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
7Q09
 
 | | URATE OXIDASE AZA-XANTHINE COMPLEX UNDER 1500 BAR OF ARGON | | Descriptor: | 8-AZAXANTHINE, ACETYL GROUP, ARGON, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
1C1M
 
 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C3L
 
 | | SUBTILISIN-CARLSBERG COMPLEXED WITH XENON (8 BAR) | | Descriptor: | CALCIUM ION, FORMIC ACID, SUBTILISIN-CARLSBERG, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
6R1Q
 
 | | murine Neuroglobin under 2 kBar of argon | | Descriptor: | ARGON, CHLORIDE ION, Neuroglobin, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2019-03-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Argon-labelling protein crystals by high pressure cooling at 2000 Bar
To Be Published
|
|
3JWE
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3J9S
 
 | |
