8VS9
 
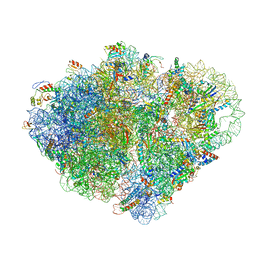 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site and deacyl-tRNA in the E site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
8VSA
 
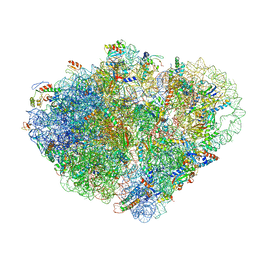 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
4HED
 
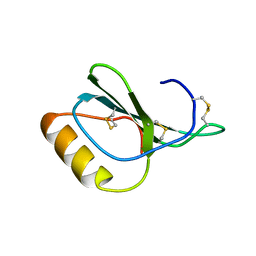 | | Zebrafish chemokine CXL1 | | Descriptor: | Uncharacterized protein | | Authors: | Rajasekaran, D, Fan, C, Meng, W, Pflugrath, J.W, Lolis, E.J. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insight into the evolution of a new chemokine family from zebrafish.
Proteins, 82, 2014
|
|
4HCS
 
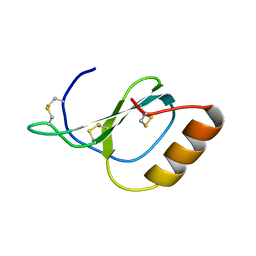 | | Structure of Novel subfamily CX chemokine solved by sulfur SAD | | Descriptor: | Uncharacterized protein | | Authors: | Rajasekaran, D, Fan, C, Meng, W, Pflugrath, J.W, Lolis, E.J. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-16 | | Last modified: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insight into the evolution of a new chemokine family from zebrafish.
Proteins, 82, 2014
|
|
3V8B
 
 | | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021
To be Published
|
|
5OLF
 
 | |
1Y9Q
 
 | |
1YBD
 
 | |
1YAV
 
 | |
1Y9I
 
 | | Crystal structure of low temperature requirement C protein from Listeria monocytogenes | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two "proton wires".
Proteins, 64, 2006
|
|
1YT8
 
 | |
2AFA
 
 | |
4ZJX
 
 | |
2OG9
 
 | |
3N52
 
 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
2OPX
 
 | |
2PGW
 
 | |
2P9B
 
 | |
3IO1
 
 | | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae | | Descriptor: | Aminobenzoyl-glutamate utilization protein, SODIUM ION, YTTRIUM (III) ION | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-13 | | Release date: | 2009-08-25 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae
To be Published
|
|
3IPI
 
 | | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei | | Descriptor: | Geranyltranstransferase, MALONIC ACID | | Authors: | Kumaran, D, Mohammed, M.B, Brown, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-08 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei
To be Published
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
3OC4
 
 | | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase, pyridine nucleotide-disulfide family, ... | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583
To be Published
|
|
3DEC
 
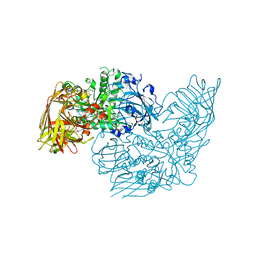 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
5IQ6
 
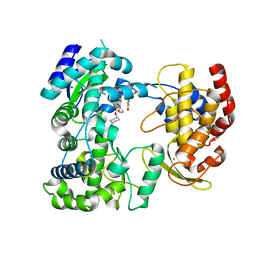 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | Authors: | Tarantino, D, Mastrangelo, E, Milani, M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
4QB5
 
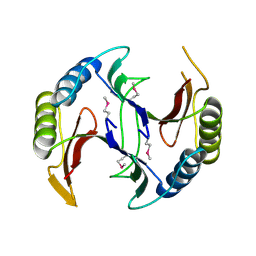 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | Descriptor: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-06 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
