3L4G
 
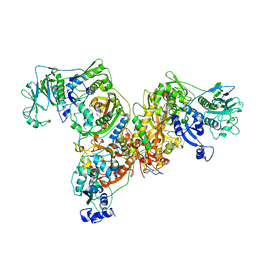 | | Crystal structure of Homo Sapiens cytoplasmic Phenylalanyl-tRNA synthetase | | Descriptor: | PHENYLALANINE, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Finarov, I, Moor, N, Kessler, N, Klipcan, L, Safro, M.G. | | Deposit date: | 2009-12-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
Structure, 18, 2010
|
|
1U6U
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1U6V
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
3HFV
 
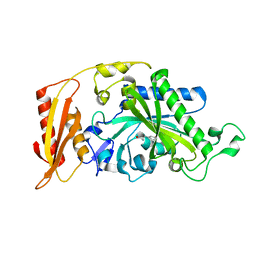 | | Crystal structure of human mitochondrial phenylalanyl-tRNA synthetase complexed with m-tyrosine | | Descriptor: | META-TYROSINE, Phenylalanyl-tRNA synthetase, mitochondrial | | Authors: | Klipcan, L, Moor, N, Kessler, N, Safro, M.G. | | Deposit date: | 2009-05-12 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HFZ
 
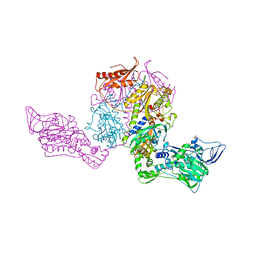 | | Crystal structure of Thermus thermophilus Phenylalanyl-tRNA synthetase complexed with m-tyrosine | | Descriptor: | META-TYROSINE, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Klipcan, L, Moor, N, Kessler, N, Safro, M.G. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3TUP
 
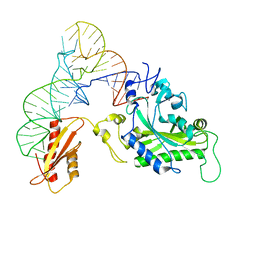 | | Crystal structure of human mitochondrial PheRS complexed with tRNAPhe in the active open state | | Descriptor: | Phenylalanyl-tRNA synthetase, mitochondrial, Thermus thermophilus tRNAPhe | | Authors: | Safro, M, Klipcan, L, Moor, N, Finarov, I, Kessler, N, Sukhanova, M. | | Deposit date: | 2011-09-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of Human Mitochondrial PheRS Complexed with tRNA(Phe) in the Active "Open" State.
J.Mol.Biol., 415, 2012
|
|
3CMQ
 
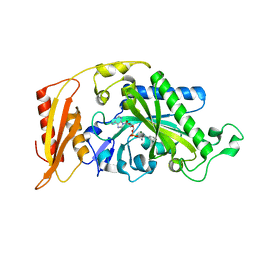 | | Crystal structure of human mitochondrial phenylalanine tRNA synthetase | | Descriptor: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], MAGNESIUM ION, Phenylalanyl-tRNA synthetase, ... | | Authors: | Klipcan, L, Levin, I.L, Kessler, N, Moor, N, Finarov, I, Safro, M. | | Deposit date: | 2008-03-24 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tRNA-Induced Conformational Activation of Human Mitochondrial Phenylalanyl-tRNA Synthetase.
Structure, 16, 2008
|
|
3PCO
 
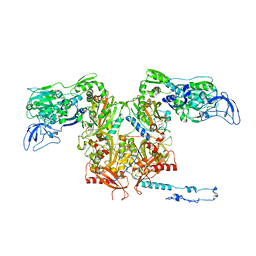 | | crystal structure of E. coli phenylalanine-tRNA synthetase complexed with phenylalanine and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHENYLALANINE, Phenylalanyl-tRNA synthetase, ... | | Authors: | Mermershtain, I, Finarov, I, Klipcan, L, Kessler, N, Rozenberg, H, Safro, M.G. | | Deposit date: | 2010-10-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Idiosyncrasy and identity in the prokaryotic phe-system: crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP.
Protein Sci., 20, 2011
|
|
2HYM
 
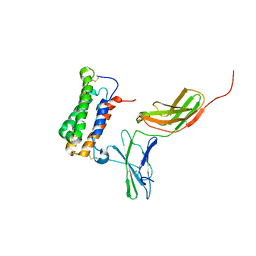 | | NMR based Docking Model of the Complex between the Human Type I Interferon Receptor and Human Interferon alpha-2 | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Quadt-Akabayov, S.R, Chill, J.H, Levy, R, Kessler, N, Anglister, J. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex
Protein Sci., 15, 2006
|
|
2AC0
 
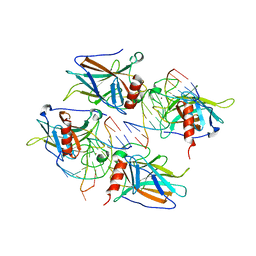 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2ADY
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex IV) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2ATA
 
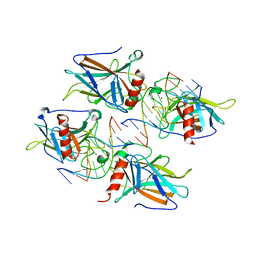 | | Structural Basis of DNA Recognition by p53 Tetramers (complex II) | | Descriptor: | 5'-D(*AP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*T)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-08-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AHI
 
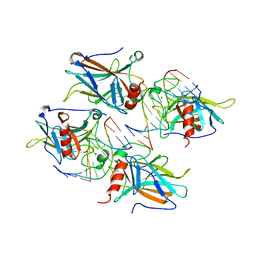 | | Structural Basis of DNA Recognition by p53 Tetramers (complex III) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1MD9
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB AND AMP | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MDF
 
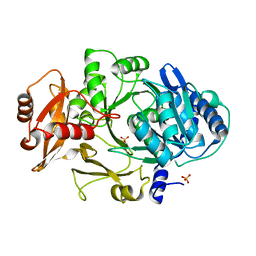 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MDB
 
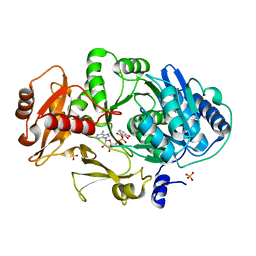 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NIZ
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1NJ0
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
2L87
 
 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | Deposit date: | 2011-01-06 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
3D05
 
 | | Human p53 core domain with hot spot mutation R249S (II) | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Suad, O, Rozenberg, H, Shimon, L.J.W, Frolow, F, Shakked, Z. | | Deposit date: | 2008-05-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations
J.Mol.Biol., 385, 2009
|
|
3D08
 
 | | Human p53 core domain with hot spot mutation R249S and second-site suppressor mutation H168R | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Suad, O, Rozenberg, H, Shimon, L.J.W, Frolow, F, Shakked, Z. | | Deposit date: | 2008-05-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations
J.Mol.Biol., 385, 2009
|
|
3D06
 
 | | Human p53 core domain with hot spot mutation R249S (I) | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Rozenberg, H, Suad, O, Shimon, L.J.W, Frolow, F, Shakked, Z. | | Deposit date: | 2008-05-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations
J.Mol.Biol., 385, 2009
|
|
3D07
 
 | |
3D09
 
 | |
3D0A
 
 | |
