1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
3CN8
 
 | |
5HL1
 
 | | Crystal structure of glutaminase C in complex with inhibitor CB-839 | | Descriptor: | 2-(pyridin-2-yl)-N-(5-{4-[6-({[3-(trifluoromethoxy)phenyl]acetyl}amino)pyridazin-3-yl]butyl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the clinically relevant glutaminase inhibitot CB-839 in complex with glutaminase C
To Be Published
|
|
1EV7
 
 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
2FM5
 
 | | Crystal structure of PDE4D2 in complex with inhibitor L-869299 | | Descriptor: | (R)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHYL)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-07 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FM0
 
 | | Crystal structure of PDE4D in complex with L-869298 | | Descriptor: | (S)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHY L)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1Z6L
 
 | |
1RSG
 
 | |
1MCT
 
 | | THE REFINED 1.6 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PORCINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR A | | Authors: | Huang, Q, Liu, S, Tang, Y. | | Deposit date: | 1992-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined 1.6 A resolution crystal structure of the complex formed between porcine beta-trypsin and MCTI-A, a trypsin inhibitor of the squash family. Detailed comparison with bovine beta-trypsin and its complex.
J.Mol.Biol., 229, 1993
|
|
1OYN
 
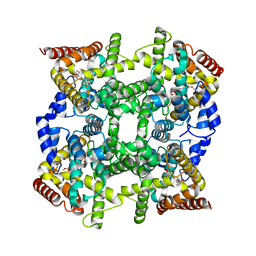 | | Crystal structure of PDE4D2 in complex with (R,S)-rolipram | | Descriptor: | ROLIPRAM, ZINC ION, cAMP-specific phosphodiesterase PDE4D2 | | Authors: | Huai, Q, Wang, H, Sun, Y, Kim, H.Y, Liu, Y, Ke, H. | | Deposit date: | 2003-04-05 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity
Structure, 11, 2003
|
|
1Q2U
 
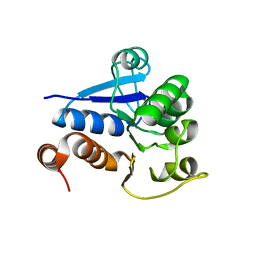 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
3BI5
 
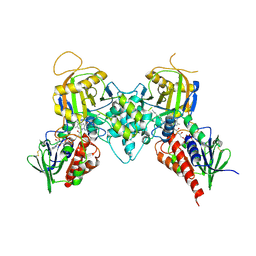 | |
3BI4
 
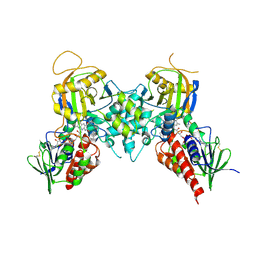 | | Crystal structures of fms1 in complex with its inhibitors | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[(1E,2Z)-but-2-en-1-ylidene]-N'-[(2E)-but-2-en-1-ylidene]butane-1,4-diamine, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-11-29 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of fms1 in complex with its inhibitors
To be Published
|
|
3BI2
 
 | |
1Q9M
 
 | | Three dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity | | Descriptor: | ROLIPRAM, ZINC ION, cAMP-specific phosphodiesterase PDE4D2 | | Authors: | Huai, Q, Wang, H, Sun, Y, Kim, H.Y, Liu, Y, Ke, H. | | Deposit date: | 2003-08-25 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity
Structure, 11, 2003
|
|
3BNM
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3R,3'R)-methylated spermine | | Descriptor: | (3R,3'R)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
3BNU
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3S,3'S)-methylated spermine | | Descriptor: | (3S,3'S)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
1PTW
 
 | |
1Q59
 
 | | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2 | | Descriptor: | Early antigen protein R | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2
J.Mol.Biol., 332, 2003
|
|
3CNP
 
 | | Crystal structure of fms1 in complex with S-N1-AcMeSpermidine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-{(1S)-3-[(4-aminobutyl)amino]-1-methylpropyl}acetamide, ... | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fms1 in complex with S-N1-AcMeSpermidine
TO BE PUBLISHED
|
|
