1EQZ
 
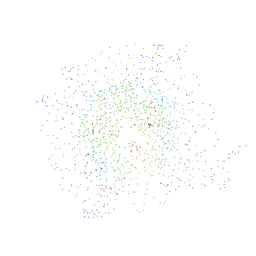 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5C6E
 
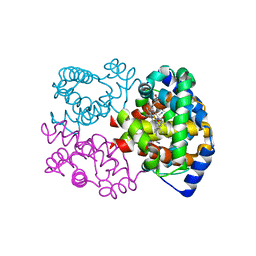 | | Joint X-ray/neutron structure of equine cyanomet hemoglobin in R state | | Descriptor: | CYANIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dajnowicz, S, Sean, S, Hanson, B.L, Fisher, S.Z, Langan, P, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2015-06-22 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the Bohr effect in hemoglobin: neutron structure of equine cyanomethemoglobin in the R state and comparison with human deoxyhemoglobin in the T state.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2GLK
 
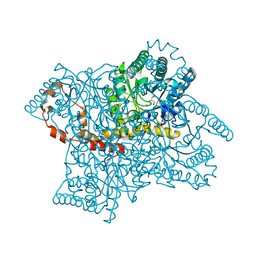 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GVE
 
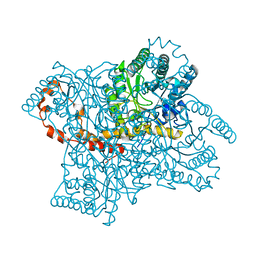 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3KGG
 
 | | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): Perdeuteration of proteins for neutron diffraction | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Tomanicek, S.J, John, H, Hanson, B.L, terjans, H.R, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): perdeuteration of proteins for neutron diffraction.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2YXP
 
 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4DVO
 
 | | Room-temperature joint X-ray/neutron structure of D-xylose isomerase in complex with 2Ni2+ and per-deuterated D-sorbitol at pH 5.9 | | Descriptor: | NICKEL (II) ION, Xylose isomerase, sorbitol | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DUO
 
 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3QYS
 
 | |
3QZA
 
 | |
4DH5
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3KBJ
 
 | |
2PKY
 
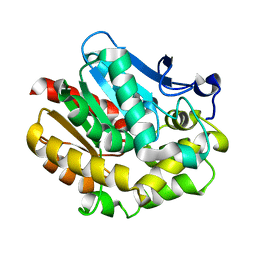 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3U7T
 
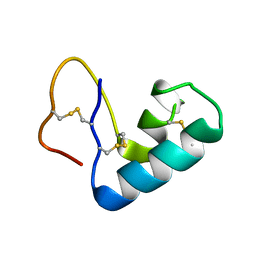 | |
4DH1
 
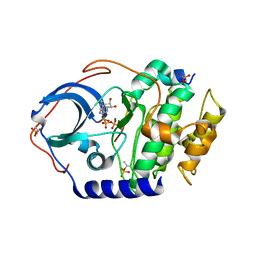 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
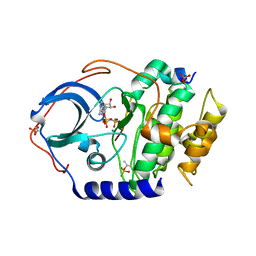 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH3
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH8
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FC1
 
 | |
3KCJ
 
 | |
2GUB
 
 | |
