8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4UI9
 
 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
5XPD
 
 | |
7YSE
 
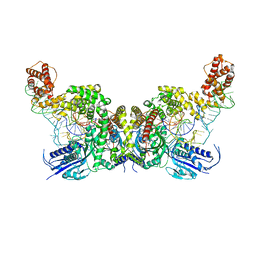 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
6C9B
 
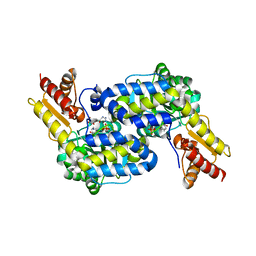 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
5BK7
 
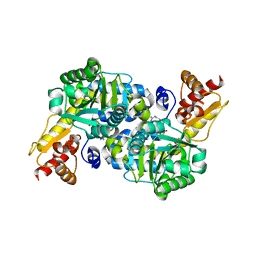 | | The structure of MppP E15A mutant soaked with the substrate L-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C92
 
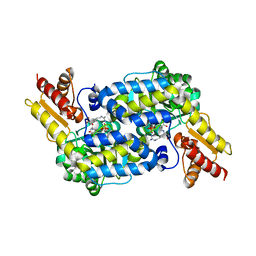 | | The structure of MppP soaked with the product 2-ketoarginine | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C8T
 
 | | The structure of MppP soaked with the substrate L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
5HOQ
 
 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5N
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5H
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 2.09 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5R
 
 | | Crystal Structure of C150S mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.31 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5I
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 2.49 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5Q
 
 | | Crystal Structure of H177A mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.13 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5K
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.69 Angstrom resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5P
 
 | | Crystal Structure Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.35 Angstrom resolution. | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5J
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C7K
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from Escherichia coli at 1.77 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5M
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 1.8 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7CLZ
 
 | |
