6L0B
 
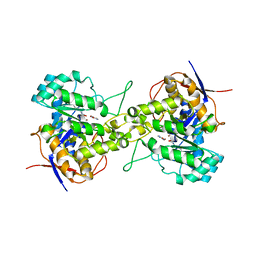 | | Crystal structure of dihydroorotase in complex with fluorouracil from Saccharomyces cerevisiae | | Descriptor: | 5-FLUOROURACIL, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0G
 
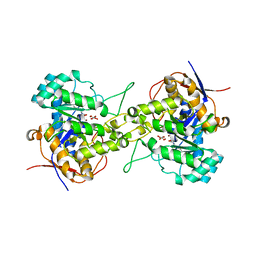 | | Crystal structure of dihydroorotase in complex with malate at pH6 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0A
 
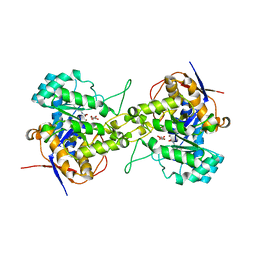 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
3K7N
 
 | | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guan, H.H, Huang, Y.W, Wu, W.G, Chen, C.J. | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins
J.Struct.Biol., 169, 2010
|
|
3K7L
 
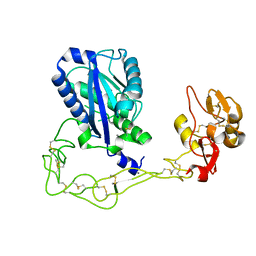 | | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Atragin, CALCIUM ION, ... | | Authors: | Guan, H.H, Wu, W.G, Chen, C.J. | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins
J.Struct.Biol., 169, 2010
|
|
5XMJ
 
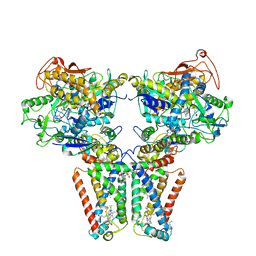 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
6L0I
 
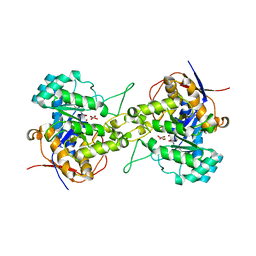 | | Crystal structure of dihydroorotase in complex with malate at pH6.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0F
 
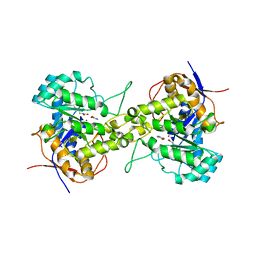 | | Crystal structure of dihydroorotase in complex with 5-Aminouracil from Saccharomyces cerevisiae | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0K
 
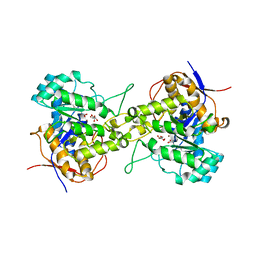 | | Crystal structure of dihydroorotase in complex with malate at pH9 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0J
 
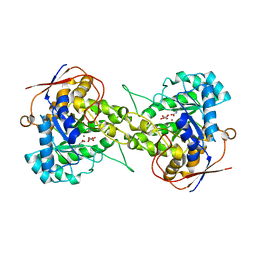 | | Crystal structure of Dihydroorotase in complex with malate at pH7.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
6L0H
 
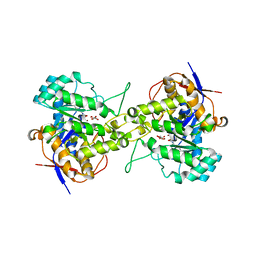 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
7CA0
 
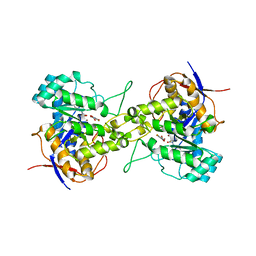 | | Crystal structure of dihydroorotase in complex with 5-fluoroorotic acid from Saccharomyces cerevisiae | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complexed Crystal Structure of Saccharomyces cerevisiae Dihydroorotase with Inhibitor 5-Fluoroorotate Reveals a New Binding Mode.
Bioinorg Chem Appl, 2021, 2021
|
|
7CA1
 
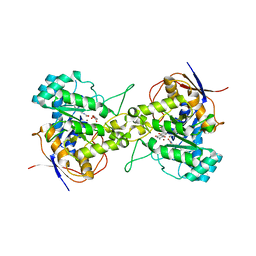 | | Crystal structure of dihydroorotase in complex with plumbagin from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 5-hydroxy-2-methylnaphthalene-1,4-dione, Dihydroorotase, ... | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis.
Int J Mol Sci, 22, 2021
|
|
8X8G
 
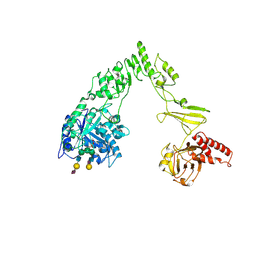 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105, in complex with oligosaccharide G2S2-oxazoline | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose, ... | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4G
 
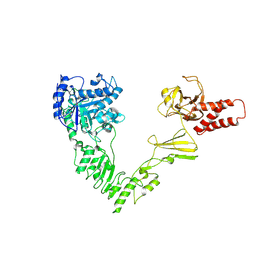 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4M
 
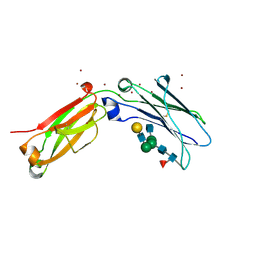 | | Crystal structure of open conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | Immunoglobulin gamma-1 heavy chain, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4I
 
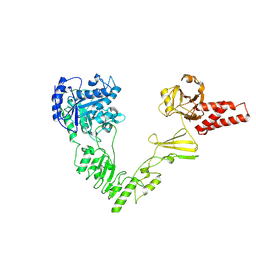 | | Crystal structure of EndoSz mutant D234M in space group P21 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4L
 
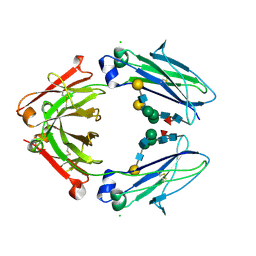 | | Crystal structure of closed conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4N
 
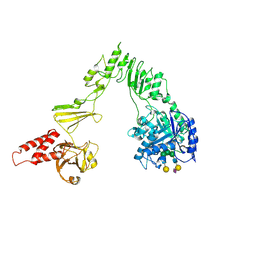 | | Crystal structure of EndoSz mutant D234M, in space group P21, in complex with oligosaccharide G2S1 | | Descriptor: | CALCIUM ION, Glycoside hydrolase, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
4WIZ
 
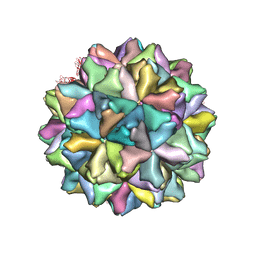 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
5YL0
 
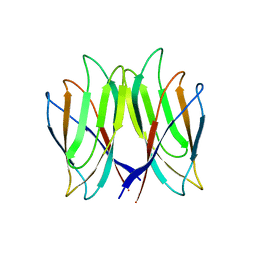 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P212121) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKX
 
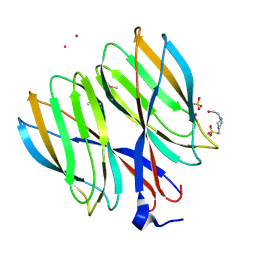 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Cd ion | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, Capsid protein, ... | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKZ
 
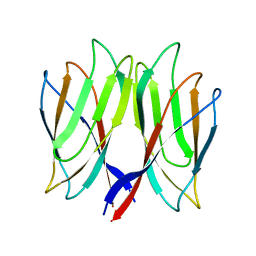 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P21) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKV
 
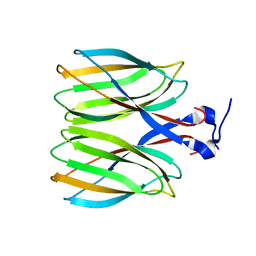 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKU
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Zn ions | | Descriptor: | Capsid protein, ZINC ION | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
