4D5S
 
 | | Structure of A49 from Vaccinia Virus Western Reserve | | Descriptor: | A49R | | Authors: | Neidel, S, Maluquer de Motes, C, Mansur, D.S, Strnadova, P, Smith, G.L, Graham, S.C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Vaccinia Virus Protein A49 is an Unexpected Member of the B-Cell Lymphoma (Bcl)-2 Protein Family
J.Biol.Chem., 290, 2015
|
|
4D5T
 
 | | Structure of N-terminally truncated A49 from Vaccinia Virus Western Reserve | | Descriptor: | PROTEIN A49R, SULFATE ION | | Authors: | Neidel, S, Maluquer de Motes, C, Mansur, D.S, Strnadova, P, Smith, G.L, Graham, S.C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Vaccinia Virus Protein A49 is an Unexpected Member of the B-Cell Lymphoma (Bcl)-2 Protein Family
J.Biol.Chem., 290, 2015
|
|
3KOF
 
 | | Crystal structure of the double mutant F178Y/R181E of E.coli transaldolase B | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Schneider, S, Gutierrez, M, Sandalova, T, Schneider, G, Clapes, P, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates.
Chembiochem, 11, 2010
|
|
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
8RC0
 
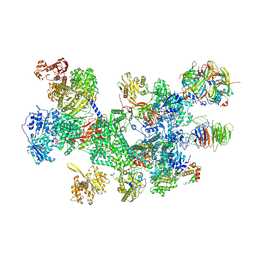 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q91
 
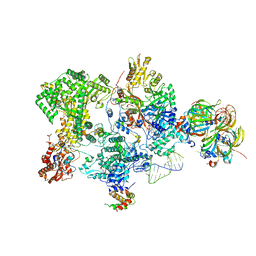 | | Structure of the human 20S U5 snRNP core | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FAI
 
 | | Cryo-EM structure of the Agrobacterium T-pilus | | Descriptor: | (7Z,19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacos-7-en-19-yl (9Z)-octadec-9-enoate, Protein virB2 | | Authors: | Kreida, S, Narita, A, Johnson, M.D, Tocheva, E.I, Das, A, Jensen, G.J, Ghosal, D. | | Deposit date: | 2022-11-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T4SS-associated T-pilus reveals stoichiometric protein-phospholipid assembly.
Structure, 31, 2023
|
|
2J0R
 
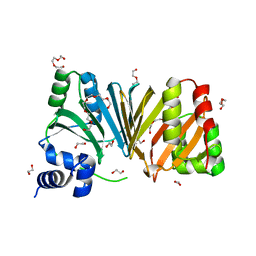 | | Structure of the haem-chaperone Proteobacteria-protein HemS | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | Authors: | Schneider, S, Sharp, K.H, Barker, P.D, Paoli, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Induced Fit Conformational Change Underlies the Binding Mechanism of the Heme Transport Proteobacteria-Protein Hems.
J.Biol.Chem., 281, 2006
|
|
2J0P
 
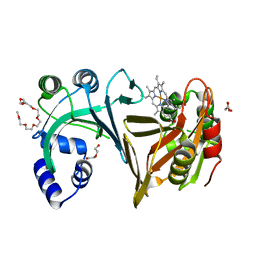 | | Structure of the haem-chaperone Proteobacteria-protein HemS | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, HEMIN TRANSPORT PROTEIN HEMS, ... | | Authors: | Schneider, S, Sharp, K.H, Barker, P.D, Paoli, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Induced Fit Conformational Change Underlies the Binding Mechanism of the Heme Transport Proteobacteria-Protein Hems.
J.Biol.Chem., 281, 2006
|
|
5HLR
 
 | | Linalool dehydratase/isomerase: Ldi-apo | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Linalool dehydratase/isomerase | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-15 | | Release date: | 2016-04-27 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
8R33
 
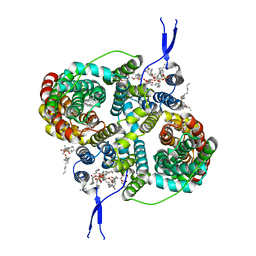 | |
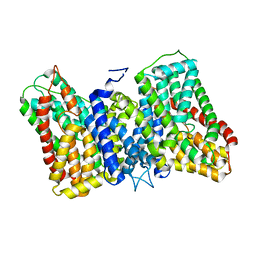
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 32, 2024
|
|
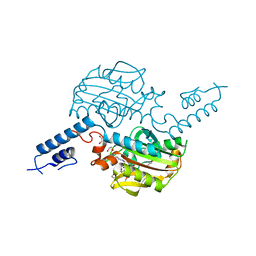
8R35
 
 | |
5HSS
 
 | | Linalool dehydratase/isomerase: Ldi with monoterpene substrate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-Myrcene, Geraniol, ... | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-26 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
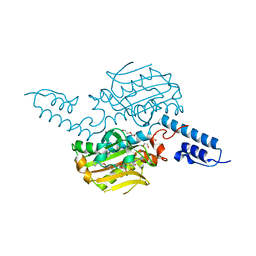
4BW9
 
 | | PylRS Y306G, Y384F, I405R mutant in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schneider, S, Vrabel, M, Gattner, M.J, Fluegel, V, Lopez-Carillo, V, Carell, T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights Into Incorporation of Norbornene Amino Acids for Click Modification of Proteins
Chem.Bio.Chem., 14, 2013
|
|
4BWA
 
 | | PylRS Y306G, Y384F, I405R mutant in complex with adenylated norbornene | | Descriptor: | 1,2-ETHANEDIOL, Adenylated Norbornene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schneider, S, Vrabel, M, Gattner, M.J, Fluegel, V, Lopez-Carillo, V, Carell, T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights Into Incorporation of Norbornene Amino Acids for Click Modification of Proteins
Chem.Bio.Chem., 14, 2013
|
|
2I5A
 
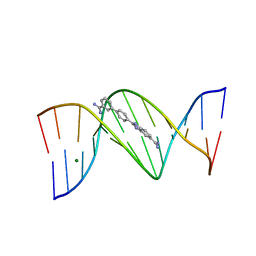 | | Crystal structure of a DB1055-D(CGCGAATTCGCG)2 complex | | Descriptor: | 2-{3'-[AMINO(IMINO)METHYL]BIPHENYL-4-YL}-1H-BENZIMIDAZOLE-5-CARBOXIMIDAMIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Neidle, S, Lee, M.P.H. | | Deposit date: | 2006-08-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a DB1055-D(CGCGAATTCGCG)2 complex
To be Published
|
|
453D
 
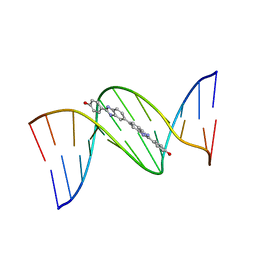 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'-BENZIMIDAZOLE COMPLEX | | Descriptor: | 4-{[4-HYDROXY-PHENYL]-1H-BENZIMIDAZOLE-5-YL}-BENZIMIDAZOLE-2-YL-[4-HYDROXY-BENZENE], DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Neidle, S. | | Deposit date: | 1999-02-18 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric bis-benzimidazoles, a new class of sequence-selective DNA-binding molecules
J.Chem.Soc.,Chem.Commun., 1999
|
|
4D5R
 
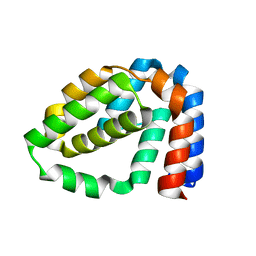 | | Structure of N-terminally truncated A49 from Vaccinia Virus Western Reserve | | Descriptor: | A49 | | Authors: | Neidel, S, Maluquer de Motes, C, Mansur, D.S, Strnadova, P, Smith, G.L, Graham, S.C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Protein A49 is an Unexpected Member of the B-Cell Lymphoma (Bcl)-2 Protein Family
J.Biol.Chem., 290, 2015
|
|
4GI2
 
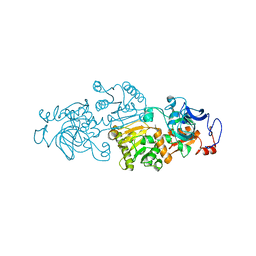 | | Crotonyl-CoA Carboxylase/Reductase | | Descriptor: | Crotonyl-CoA carboxylase/reductase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weidenweber, S, Erb, T.J, Ermler, U. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crotonyl-CoA Carboxylase/Reductase
To be Published
|
|
6YX5
 
 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
5LCL
 
 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH C8-aminofluorene- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, GCTCTAC(8AF)TCATCA, ... | | Authors: | Schneider, S, Carell, T, Ebert, C, Simon, N. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
4CIS
 
 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
5LCM
 
 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH N2-acetylaminonaphtyl- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*TP*AP*CP*(AAN)P*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, ... | | Authors: | Schneider, S, Ebert, C, Simon, N, Carell, T. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5LIR
 
 | |
