5H1T
 
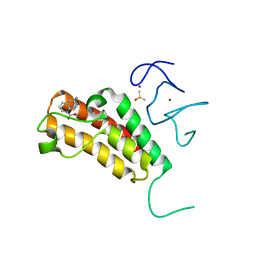 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
5GNK
 
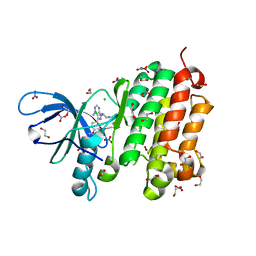 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
4R7M
 
 | |
4R5V
 
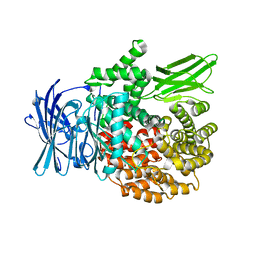 | |
3DG7
 
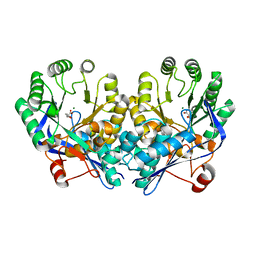 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
4R5X
 
 | |
5GTY
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
5H1V
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 6 | | Descriptor: | 2-Hydrazino-1,3-benzothiazole-6-carbohydrazide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
8T1Y
 
 | |
8T27
 
 | |
8T1Z
 
 | |
8T24
 
 | |
8T26
 
 | |
7QVJ
 
 | | ESTROGEN RECEPTOR ALPHA IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 2,2-bis(fluoranyl)-3-[(1~{R},3~{R})-1-[6-fluoranyl-3-[2-(3-fluoranylpropylamino)ethoxy]-2-methyl-phenyl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]propan-1-ol, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
7QVL
 
 | | OESTROGEN RECEPTOR LIGAND BINDING DOMAIN IN COMPLEX WITH COMPOUND 38 | | Descriptor: | (2~{R})-3-[(1~{R},3~{R})-1-[5-fluoranyl-2-[2-(3-fluoranylpropylamino)ethoxy]-3-methyl-pyridin-4-yl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]-2-methyl-propanoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
4R76
 
 | |
7CM9
 
 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
4R5T
 
 | |
4R6T
 
 | |
7R4U
 
 | | Apoform of FtrA/P19 from Rubrivivax gelatinosus | | Descriptor: | FtrA-P19, GLYCEROL, SODIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Plancqueel, S. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | New insights into the mechanism of iron transport through the bacterial Ftr system present in pathogens.
Febs J., 289, 2022
|
|
7R3S
 
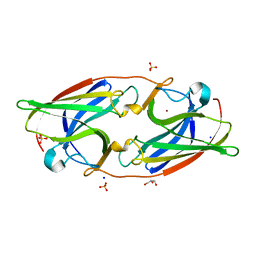 | | FtrA/P19 of Rubrivivax gelatinosus in complex with Ni | | Descriptor: | FtrA-P19 protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2022-02-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | New insights into the mechanism of iron transport through the bacterial Ftr system present in pathogens.
Febs J., 289, 2022
|
|
7R5G
 
 | |
7R4Z
 
 | |
7R3P
 
 | |
