6M5U
 
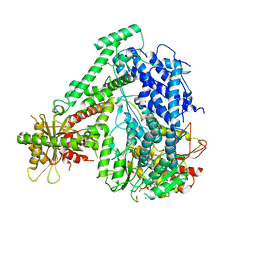 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
7CAC
 
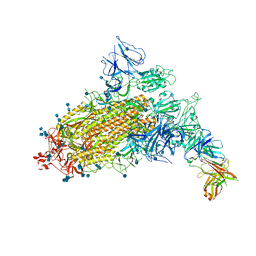 | | SARS-CoV-2 S trimer with one RBD in the open state and complexed with one H014 Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
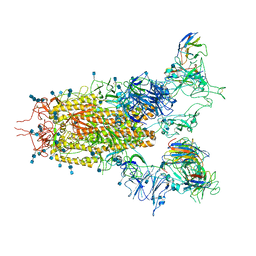 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CMK
 
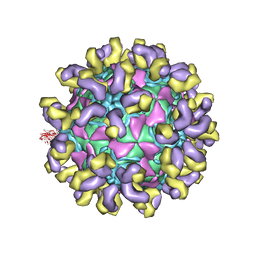 | | E30 E-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, VP1, ... | | Authors: | Wang, K, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7CAB
 
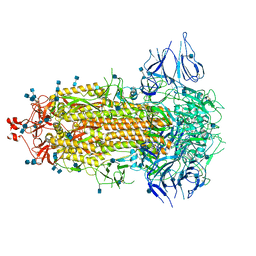 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7C9Z
 
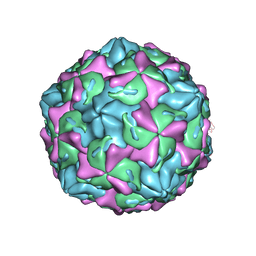 | | Coxsackievirus B1 F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Feng, R, Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9X
 
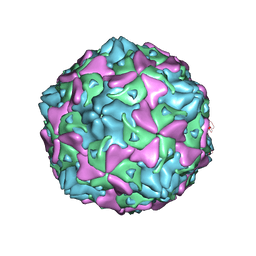 | | Echovirus 3 F-particle | | Descriptor: | MYRISTIC ACID, SPHINGOSINE, VP1, ... | | Authors: | Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
6O6F
 
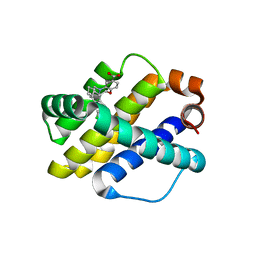 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5'-chloro-5-(cyclobutylmethyl)-2',3',4,5-tetrahydro-2H-spiro[1,5-benzoxazepine-3,1'-indene]-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
7C9Y
 
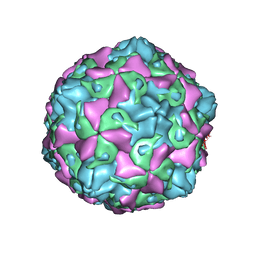 | | Coxsackievirus B5 (CVB5) F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
6DUB
 
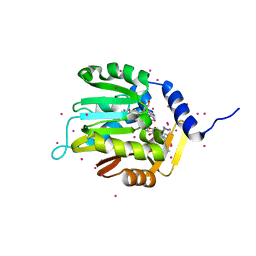 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
1SDA
 
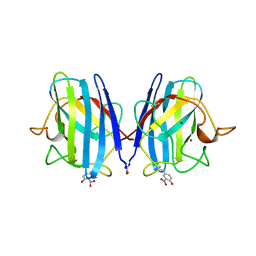 | | CRYSTAL STRUCTURE OF PEROXYNITRITE-MODIFIED BOVINE CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Smith, C.D, Carson, M, Van Der Woerd, M, Chen, J, Ischiropoulos, H, Beckman, J.S. | | Deposit date: | 1993-01-13 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of peroxynitrite-modified bovine Cu,Zn superoxide dismutase.
Arch.Biochem.Biophys., 299, 1992
|
|
7XUX
 
 | |
7JS6
 
 | | Solution NMR structure of des-citrulassin F | | Descriptor: | des-citrulassin F | | Authors: | Harris, L.A, Mitchell, D.A. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Reactivity-Based Screening for Citrulline-Containing Natural Products Reveals a Family of Bacterial Peptidyl Arginine Deiminases.
Acs Chem.Biol., 15, 2020
|
|
7RC3
 
 | | Aeronamide N-methyltransferase, AerE (Y137F) | | Descriptor: | ASPARTIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC2
 
 | | Aeronamide N-methyltransferase, AerE | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC4
 
 | | Aeronamide N-methyltransferase, AerE (D141A) | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Methyltransferase family protein, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC6
 
 | | Aeronamide N-methyltransferase, AerE, bound to modified peptide substrate, AerA-DL,34 | | Descriptor: | Aeronamide A peptide, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC5
 
 | | Aeronamide N-methyltransferase, AerE (N231A) | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5CDZ
 
 | | Crystal structure of conserpin in the latent state | | Descriptor: | Conserpin in the latent state, GLYCEROL | | Authors: | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
5CE0
 
 | |
5CDX
 
 | |
5HPY
 
 | | Crystal Structure of RhoA.GDP.MgF3-in complex with human Myosin 9b RhoGAP domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRIFLUOROMAGNESATE, ... | | Authors: | Yi, F.S, Ren, J.Q, Feng, W. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncanonical Myo9b-RhoGAP Accelerates RhoA GTP Hydrolysis by a Dual-Arginine-Finger Mechanism
J.Mol.Biol., 428, 2016
|
|
5EPO
 
 | | The three-dimensional structure of Clostridium absonum 7alpha-hydroxysteroid dehydrogenase | | Descriptor: | 7-alpha-hydroxysteroid deydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lou, D, Wang, B, Wang, F. | | Deposit date: | 2015-11-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Clostridium absonum 7 alpha-hydroxysteroid dehydrogenase: new insights into the conserved arginines for NADP(H) recognition
Sci Rep, 6, 2016
|
|
7SLI
 
 | | Engineered sperm whale myoglobin-based carbene transferase MbBTIC-C2 | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Jenkins, J.L, Vargas, D, Ren, X, Fasan, R. | | Deposit date: | 2021-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic strategy for the construction of sp 3 -rich polycyclic compounds from directed evolution and computational modelling.
Nat.Chem., 16, 2024
|
|
