3LRS
 
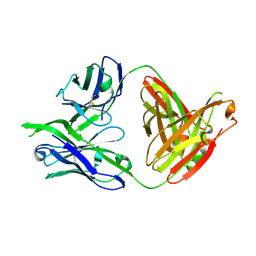 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
5TDL
 
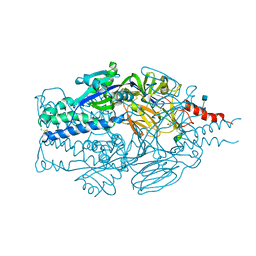 | |
5TDG
 
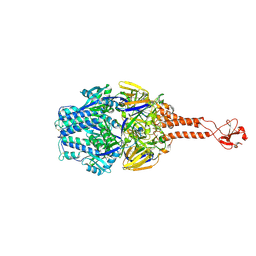 | |
7JZI
 
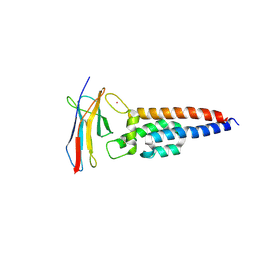 | |
7JZ4
 
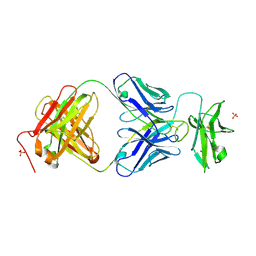 | |
7JZK
 
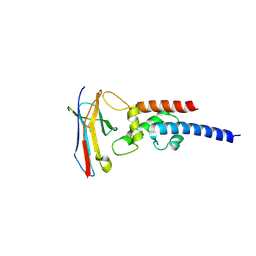 | |
7JZ1
 
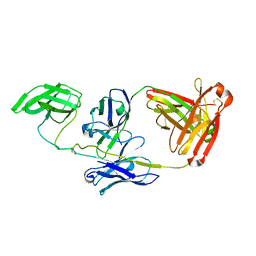 | |
5UTY
 
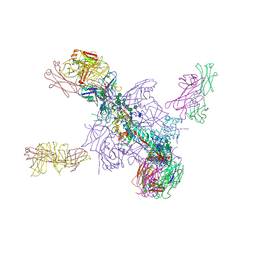 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
7BH4
 
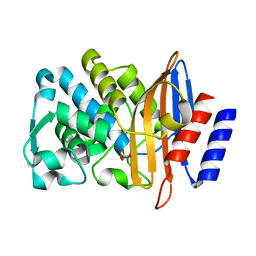 | | XFEL structure of apo CTX-M-15 after mixing for 0.7 sec with ertapenem using a piezoelectric injector (PolyPico) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHN
 
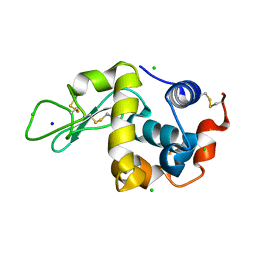 | |
7BH5
 
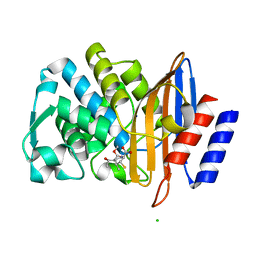 | | XFEL structure of the ertapenem-derived CTX-M-15 acylenzyme after mixing for 2 sec using a piezoelectric injector (PolyPico) | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHM
 
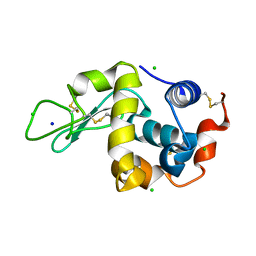 | |
7BH7
 
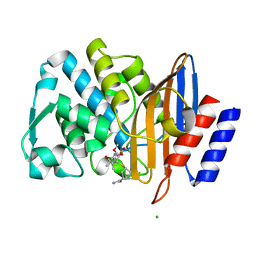 | | Room temperature, serial X-ray structure of the ertapenem-derived acylenzyme of CTX-M-15 (10 min soak) collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH6
 
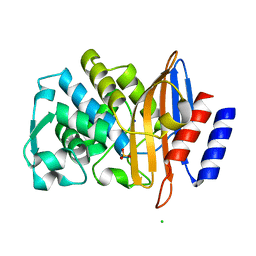 | | Room temperature, serial X-ray structure of CTX-M-15 collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHK
 
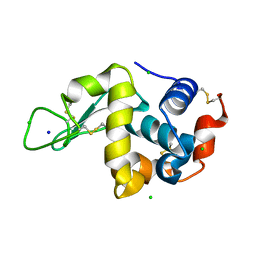 | |
7BH3
 
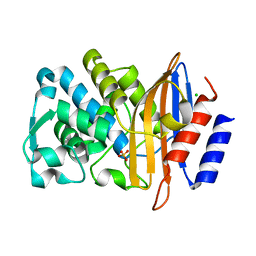 | | XFEL structure of CTX-M-15 resting state | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHL
 
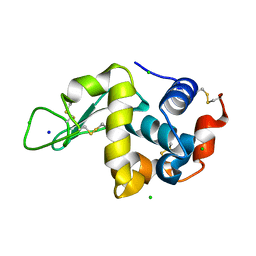 | |
3TIH
 
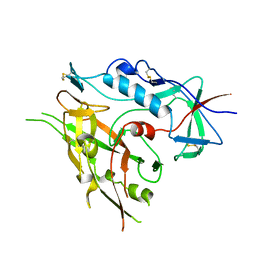 | |
3U1S
 
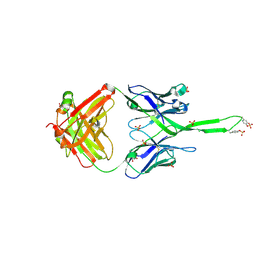 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
6XSK
 
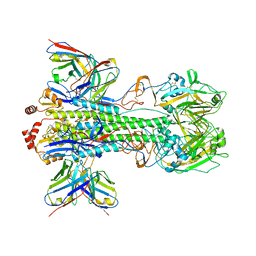 | | Cryo-EM Structure of Vaccine-Elicited Rhesus Antibody 789-203-3C12 in Complex with Stabilized SI06 (A/Solomon Islands/3/06) Influenza Hemagglutinin Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 789-203-3C12 Fab Heavy Chain, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses.
Immunity, 55, 2022
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGR
 
 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGQ
 
 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGS
 
 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TCL
 
 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
