7VIC
 
 | | The crystal structure of SARS-CoV-2 3C-like protease in complex with a traditional Chinese Medicine Inhibitors | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase | | Authors: | Zhong, B, Chen, B, Zhou, H, Sun, L. | | Deposit date: | 2021-09-26 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oridonin Inhibits SARS-CoV-2 by Targeting Its 3C-Like Protease.
Small Sci, 2, 2022
|
|
7WOB
 
 | | SARS-CoV-2 Spike in complex with IgG 553-60 (2-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOG
 
 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | Descriptor: | 553-49 VH, 553-49 VL, Spike protein S1 | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO4
 
 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 dimer trimer ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO7
 
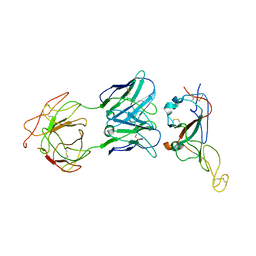 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ2
 
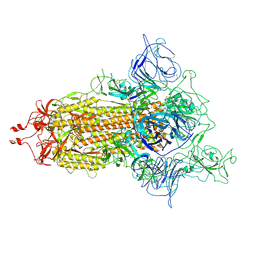 | | SARS-CoV-2 (D614G) Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO5
 
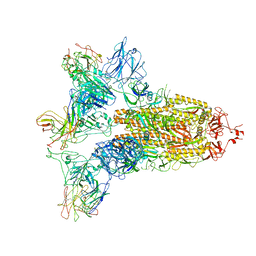 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOC
 
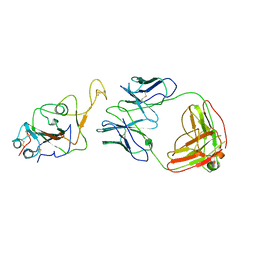 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOA
 
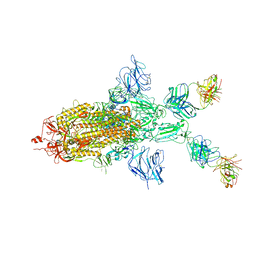 | | SARS-CoV-2 Spike in complex with IgG 553-60 (1-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ1
 
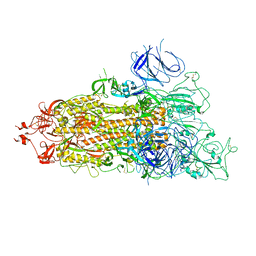 | | SARS-CoV-2 Omicron Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
8Z6X
 
 | |
8Z6R
 
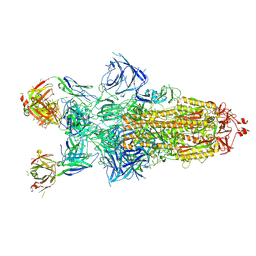 | | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1. | | Descriptor: | CYFN1006-1 heavy chain, CYFN1006-1 light chain, Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Sun, L. | | Deposit date: | 2024-04-19 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1.
To Be Published
|
|
8Z6W
 
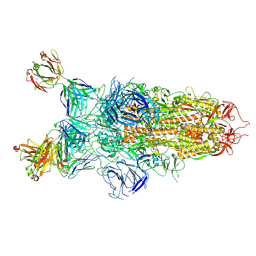 | |
8Z6U
 
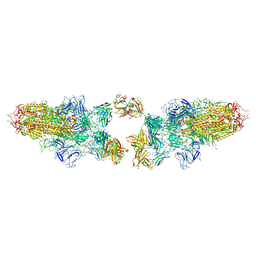 | |
8Z6S
 
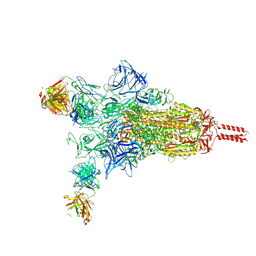 | |
8Z6Q
 
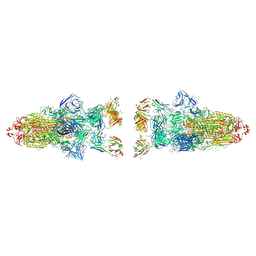 | |
8Y6V
 
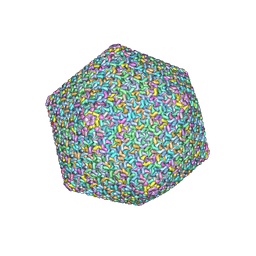 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | Descriptor: | gp119, gp120, gp162, ... | | Authors: | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
8Z6T
 
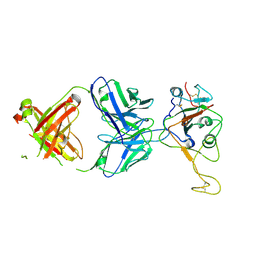 | |
8ZPX
 
 | |
8ZQ4
 
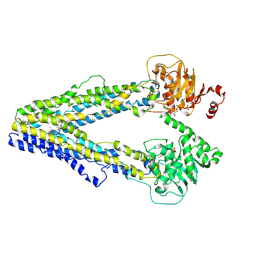 | | Structure of the Arabidopsis ABCB1 EQ mutant in the BL plus ATP bound state | | Descriptor: | ABC transporter B family member 1, ADENOSINE-5'-TRIPHOSPHATE, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2024-05-31 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into brassinosteroid export mediated by the Arabidopsis ABC transporter ABCB1.
Plant Commun., 6, 2025
|
|
8ZPZ
 
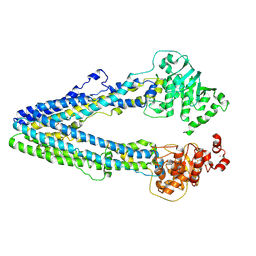 | | Structure of the wild-type Arabidopsis ABCB1 in the brassinolide-bound state | | Descriptor: | ABC transporter B family member 1, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2024-05-31 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into brassinosteroid export mediated by the Arabidopsis ABC transporter ABCB1.
Plant Commun., 6, 2025
|
|
7E5O
 
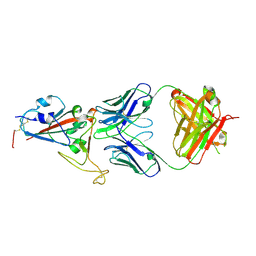 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NJ6
 
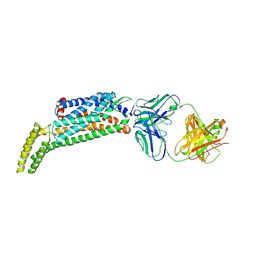 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
