9JHX
 
 | | Versatile Aromatic Prenyltransferase auraA mutant-Y207A in complex with DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, Versatile Aromatic Prenyltransferase auraA | | Authors: | Li, D, Zhang, Y, Wang, W, Wang, P. | | Deposit date: | 2024-09-10 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Characterization and structural analysis of a versatile aromatic prenyltransferase for imidazole-containing diketopiperazines.
Nat Commun, 16, 2025
|
|
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
8X8O
 
 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) in 5 mM EDTA | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
8X8M
 
 | | Cryo-EM structure of a bacteriophage tail- spike protein against Klebsiella pneumoniae K64,ORF41(K64-ORF41) | | Descriptor: | Probable tail spike protein | | Authors: | Xie, Y, Huang, T, Zhang, Z, Tao, X. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Int.J.Biol.Macromol., 265, 2024
|
|
8Y9E
 
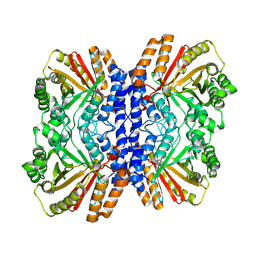 | |
8Y9D
 
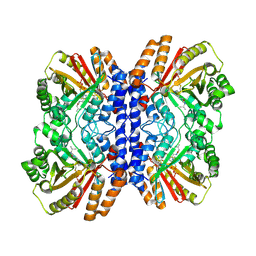 | | Versatile Aromatic Prenyltransferase auraA in complex with DMAPP and cyclo-(L-Val-L-His) | | Descriptor: | (3~{S},6~{S})-3-(1~{H}-imidazol-4-ylmethyl)-6-propan-2-yl-piperazine-2,5-dione, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Versatile Aromatic Prenyltransferase auraA | | Authors: | Li, D, Zhang, Y, Wang, W, Wang, P. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Characterization and structural analysis of a versatile aromatic prenyltransferase for imidazole-containing diketopiperazines.
Nat Commun, 16, 2025
|
|
8Y9G
 
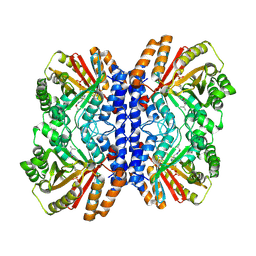 | | Versatile Aromatic Prenyltransferase auraA in complex with DMSPP and cyclo-(L-Val-DH-His) | | Descriptor: | (3~{Z},6~{S})-3-(1~{H}-imidazol-4-ylmethylidene)-6-propan-2-yl-piperazine-2,5-dione, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Versatile Aromatic Prenyltransferase auraA | | Authors: | Li, D, Zhang, Y, Wang, W, Wang, P. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Characterization and structural analysis of a versatile aromatic prenyltransferase for imidazole-containing diketopiperazines.
Nat Commun, 16, 2025
|
|
6II4
 
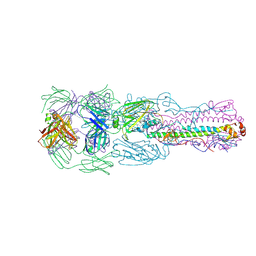 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L4A-14 | | Descriptor: | Heavy chain of L4A-14 Fab, Hemagglutinin, Light chain of L4A-14 Fab | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6II9
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L3A-44 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of L3A-44 Fab, Hemagglutinin, ... | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
6II8
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L4B-18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of L4B-18 Fab, Hemagglutinin, ... | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
7DXI
 
 | |
6Y3X
 
 | |
8RUI
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and intronistat B after 1h soaking | | Descriptor: | Domains 1-5, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RUH
 
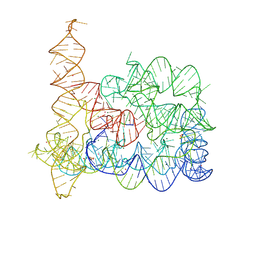 | |
8RUJ
 
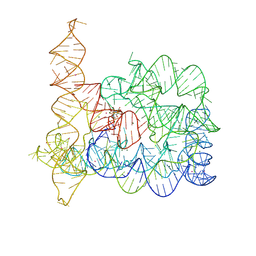 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and ARN25850 after 1h soaking | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RUL
 
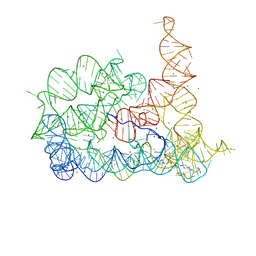 | |
8RUM
 
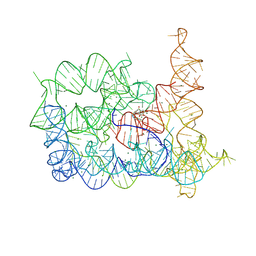 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and intronistat B | | Descriptor: | Domains 1-5, MAGNESIUM ION, ~{N}-(2-pyrrolidin-1-ylethyl)-2-[3,4,5-tris(oxidanyl)phenyl]carbonyl-1-benzofuran-5-carboxamide | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7NG2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain in apo form | | Descriptor: | ISOPROPYL ALCOHOL, Zinc finger (CCCH type) motif-containing protein | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
7NJC
 
 | |
7NH2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain bound to m6A | | Descriptor: | TETRAETHYLENE GLYCOL, Zinc finger (CCCH type) motif-containing protein, ~{N},9-dimethylpurin-6-amine | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
8RUK
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Na+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (4.81 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RUN
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Domains 1-5, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
6ZFE
 
 | |
