8J9G
 
 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J8H
 
 | | SPARTA monomer bound with guide-target, state 2 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J84
 
 | | Short ago complexed with TIR-APAZ | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Guo, L.J, Huang, P.P, Li, Z.X, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-04-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J9P
 
 | | SPARTA dimer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
9IW3
 
 | | Cryo-EM structure of Lactobacillus casei DdmE bound with guide and target | | Descriptor: | DNA (25-MER), DNA (5'-D(P*CP*TP*TP*GP*AP*TP*AP*CP*GP*AP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*GP*CP*TP*CP*TP*AP*AP*TP*CP*T)-3'), ... | | Authors: | Huang, P.P, Chen, M.R, Xiao, Y.B. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | The mechanism of bacterial defense system DdmDE from Lactobacillus casei.
Cell Res., 34, 2024
|
|
9IX4
 
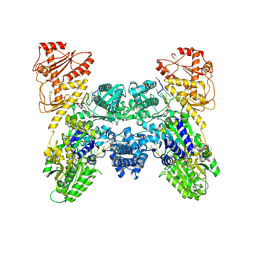 | | Cryo-EM structure of Lactobacillus casei DdmD dimer bound with DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*TP*GP*AP*GP*TP*AP*TP*AP*TP*CP*C)-3'), ... | | Authors: | Huang, P.P, Chen, M.R, Xiao, Y.B. | | Deposit date: | 2024-07-26 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The mechanism of bacterial defense system DdmDE from Lactobacillus casei.
Cell Res., 34, 2024
|
|
9IXM
 
 | | Cryo-EM structure of Lactobacillus casei DdmDE bound with DNA | | Descriptor: | DNA (30-MER), DNA (5'-D(P*TP*GP*AP*CP*GP*GP*CP*TP*CP*TP*AP*AP*TP*CP*T)-3'), DdmD, ... | | Authors: | Huang, P.P, Chen, M.R, Xiao, Y.B. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | The mechanism of bacterial defense system DdmDE from Lactobacillus casei.
Cell Res., 34, 2024
|
|
3H00
 
 | |
3V5Q
 
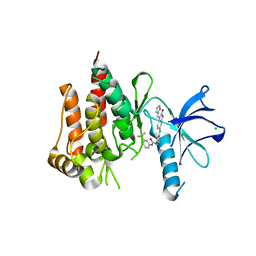 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
3G7A
 
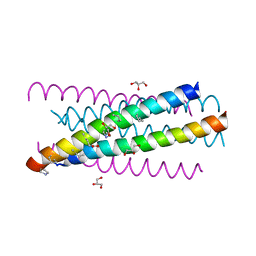 | | HIV gp41 six-helix bundle composed of a chimeric alpha+alpha/beta-peptide analogue of the CHR domain in complex with an NHR domain alpha-peptide | | Descriptor: | ACETYL GROUP, Chimeric alpha+alpha/beta-peptide analogue of the HIV gp41 CHR domain, Envelope glycoprotein gp160, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7XK8
 
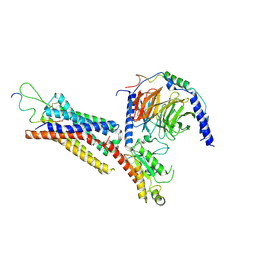 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
8Z3K
 
 | | The structure of type III CRISPR-associated deaminase in complex 2cA6-2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z40
 
 | |
8Z3R
 
 | | The structure of type III CRISPR-associated deaminase in complex cA4 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Adenosine deaminase domain-containing protein, ZINC ION | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z3P
 
 | | The structure of type III CRISPR-associated deaminase in complex cA6 and ATP, fully activated | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
7XMR
 
 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
8ZVX
 
 | | Crystal structure of snFPITE-n2 | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Chen, X, Yang, H. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
8ZVS
 
 | | Crystal structure of snFPITE-n1 | | Descriptor: | SULFATE ION, phenylmethanesulfonic acid, snFPITE-n1 | | Authors: | Chen, X, Yang, H, Hu, Y.L. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
3H01
 
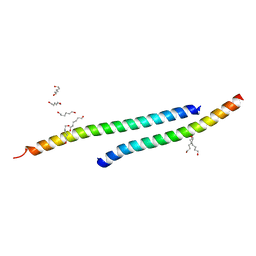 | |
3GWO
 
 | | Structure of the C-terminal Domain of a Putative HIV-1 gp41 Fusion Intermediate | | Descriptor: | Envelope glycoprotein gp160, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2009-04-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Role of a putative gp41 dimerization domain in human immunodeficiency virus type 1 membrane fusion.
J.Virol., 84, 2010
|
|
3CK4
 
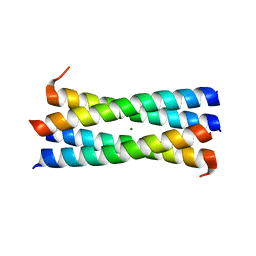 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, MAGNESIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
4FFV
 
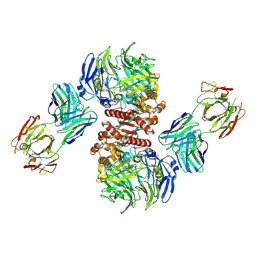 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4OCH
 
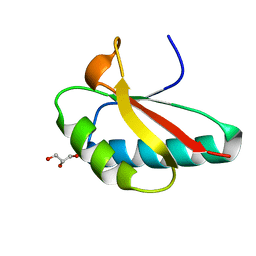 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
