6QCC
 
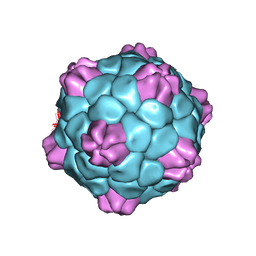 | | Cryo-EM Atomic Structure of Broad Bean Stain Virus (BBSV) | | Descriptor: | Large coat-protein subunit, Small coat-protein subunit | | Authors: | Lecorre, F, Lai Jee Him, J, Blanc, S, Zeddam, J.-L, Trapani, S, Bron, P. | | Deposit date: | 2018-12-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The cryo-electron microscopy structure of Broad Bean Stain Virus suggests a common capsid assembly mechanism among comoviruses.
Virology, 530, 2019
|
|
4ISC
 
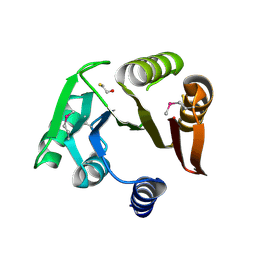 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
3GLV
 
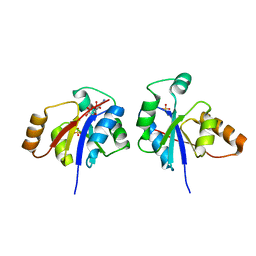 | | Crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lipopolysaccharide core biosynthesis protein, SULFATE ION | | Authors: | Zhang, R, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1
To be Published
|
|
3H04
 
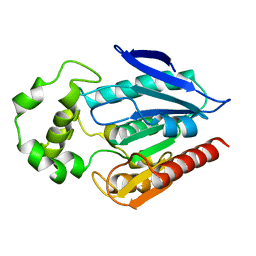 | | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | uncharacterized protein | | Authors: | Zhang, R, Tesar, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
3HH1
 
 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
4KQC
 
 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
3FRM
 
 | | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
4KND
 
 | | Thioredoxin from Anaeromyxobacter dehalogenans. | | Descriptor: | Thioredoxin | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Clancy, S, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thioredoxin from Anaeromyxobacter dehalogenans.
To be Published
|
|
3FIX
 
 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
6S44
 
 | |
3GBY
 
 | | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Uncharacterized protein CT1051 | | Authors: | Fan, Y, Chang, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum
To be Published
|
|
3FXW
 
 | | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3/inhibitor 2 complex | | Descriptor: | 2-[2-(2-FLUOROPHENYL)PYRIDIN-4-YL]-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Cheng, R.K.Y, Barker, J, Palan, S, Felicetti, B, Whittaker, M, Hesterkamp, T. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3/inhibitor 2 complex
To be Published
|
|
3G8W
 
 | | Crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, Lactococcal prophage ps3 protein 05 | | Authors: | Tan, K, Sather, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3HZ1
 
 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
3HYY
 
 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
3IC9
 
 | | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, dihydrolipoamide dehydrogenase | | Authors: | Tan, K, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H.
To be Published
|
|
4LZK
 
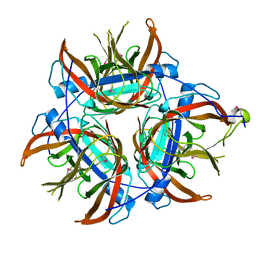 | |
3IX7
 
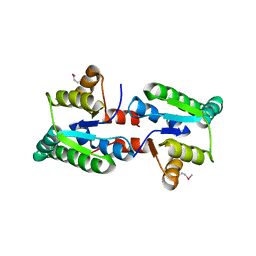 | |
3FLE
 
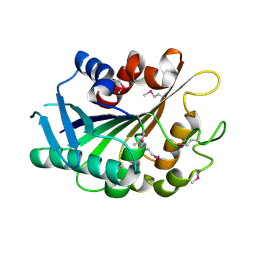 | | SE_1780 protein of unknown function from Staphylococcus epidermidis. | | Descriptor: | SE_1780 protein | | Authors: | Osipiuk, J, Hatzos, C, Clancy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray crystal structure of SE_1780 protein of unknown function from Staphylococcus epidermidis.
To be Published
|
|
3FHR
 
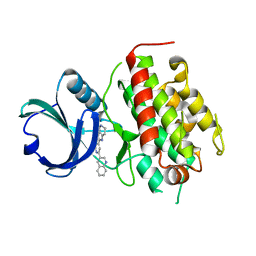 | | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3 (MK3)-inhibitor complex | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Cheng, R.K.Y, Barker, J, Palan, S, Felicetti, B, Whittaker, M, Hesterkamp, T. | | Deposit date: | 2008-12-10 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of human Mapkap kinase 3 in complex with a high affinity ligand
Protein Sci., 19, 2010
|
|
3GAA
 
 | |
4M7O
 
 | |
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
3HYZ
 
 | | Crystal structure of Hsp90 with fragment 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragment 42-C03
TO BE PUBLISHED
|
|
