3QQU
 
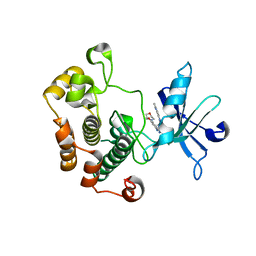 | | Cocrystal structure of unphosphorylated igf with pyrimidine 8 | | Descriptor: | Insulin-like growth factor 1 receptor, N~2~-[3-methoxy-4-(morpholin-4-yl)phenyl]-N~4~-(quinolin-3-yl)pyrimidine-2,4-diamine | | Authors: | Huang, X. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5XUO
 
 | |
5ZFZ
 
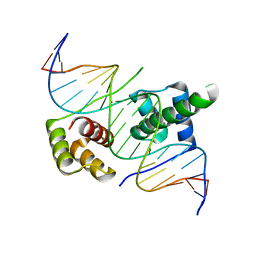 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFW
 
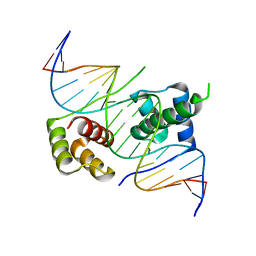 | | Crystal structure of human DUX4 homeodomains bound to A11G DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*GP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*CP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
5ZFY
 
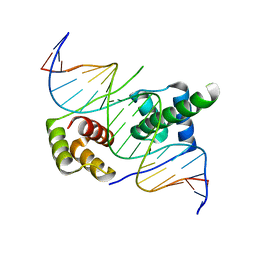 | | Crystal structure of human DUX4 homeodomains bound to A12C DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*CP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*GP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5Z6Z
 
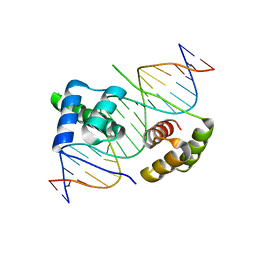 | | Crystal structure of human DUX4 homeodomains bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-01-26 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6A0Z
 
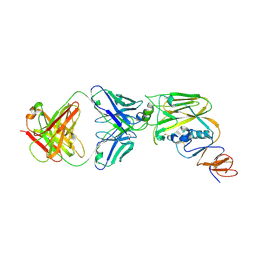 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
5ZVV
 
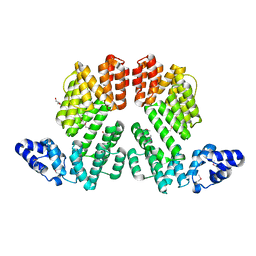 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
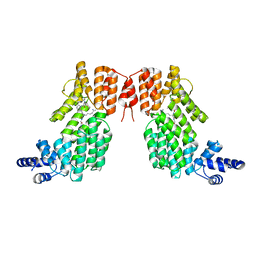 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
6A0X
 
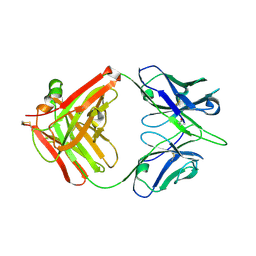 | | Crystal structure of broadly neutralizing antibody 13D4 | | Descriptor: | Antibody 13D4, Fab Heavy Chain, Fab Light Chain | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
6AJ2
 
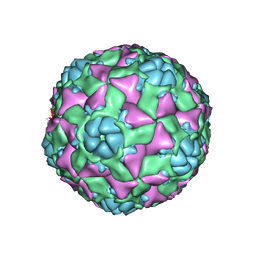 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ0
 
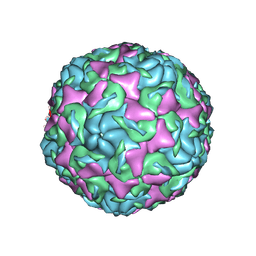 | | The structure of Enterovirus D68 mature virion | | Descriptor: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ9
 
 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
5ZW5
 
 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
